Note
Go to the end to download the full example code.
Evaluate Photon Plan Robustness#
author: OpenTPS team
This example shows how to evaluate a photon plan robustness using OpenTPS.
running time: ~ 15 minutes
Setting up the environment in google collab#
import sys
if "google.colab" in sys.modules:
from IPython import get_ipython
get_ipython().system('git clone https://gitlab.com/openmcsquare/opentps.git')
get_ipython().system('pip install ./opentps')
get_ipython().system('pip install scipy==1.10.1')
import opentps
imports
import os
import logging
import numpy as np
from matplotlib import pyplot as plt
import the needed opentps.core packages
from opentps.core.data.images import CTImage
from opentps.core.data.images import ROIMask
from opentps.core.data import Patient
from opentps.core.io import mcsquareIO
from opentps.core.io.scannerReader import readScanner
from opentps.core.io.serializedObjectIO import saveRTPlan, loadRTPlan
from opentps.core.processing.doseCalculation.doseCalculationConfig import DoseCalculationConfig
from opentps.core.processing.planEvaluation.robustnessEvaluation import RobustnessEvalPhoton
from opentps.core.processing.doseCalculation.photons.cccDoseCalculator import CCCDoseCalculator
from opentps.core.data.plan import PhotonPlanDesign
logger = logging.getLogger(__name__)
Output path#
output_path = os.path.join(os.getcwd(), 'Photon_Robust_Output_Example')
if not os.path.exists(output_path):
os.makedirs(output_path)
logger.info('Files will be stored in {}'.format(output_path))
CT calibration and BDL#
# Generic example: box of water with squared target
ctCalibration = readScanner(DoseCalculationConfig().scannerFolder)
bdl = mcsquareIO.readBDL(DoseCalculationConfig().bdlFile)
CT and ROI creation#
patient = Patient()
patient.name = 'Patient'
ctSize = 150
ct = CTImage()
ct.name = 'CT'
ct.patient = patient
huAir = -1024.
huWater = 0
data = huAir * np.ones((ctSize, ctSize, ctSize))
data[:, 50:, :] = huWater
ct.imageArray = data
roi = ROIMask()
roi.patient = patient
roi.name = 'TV'
roi.color = (255, 0, 0) # red
data = np.zeros((ctSize, ctSize, ctSize)).astype(bool)
data[100:120, 100:120, 100:120] = True
roi.imageArray = data
Create output folder
if not os.path.isdir(output_path):
os.mkdir(output_path)
# Design plan
beamNames = ["Beam1", "Beam2"]
gantryAngles = [0., 90.]
couchAngles = [0.,0]
Configure MCsquare
ccc = CCCDoseCalculator(batchSize= 30)
ccc.ctCalibration = readScanner(DoseCalculationConfig().scannerFolder)
Load / Generate new plan
plan_file = os.path.join(output_path, "Plan_Photon_WaterPhantom_notCropped_optimized.tps")
# plan_file = os.path.join(output_path, "Plan_Photon_WaterPhantom_cropped_optimized.tps")
if os.path.isfile(plan_file):
plan = loadRTPlan(plan_file, 'photon')
logger.info('Plan loaded')
else:
planDesign = PhotonPlanDesign()
planDesign.ct = ct
planDesign.targetMask = roi
planDesign.isocenterPosition_mm = None # None take the center of mass of the target
planDesign.gantryAngles = gantryAngles
planDesign.couchAngles = couchAngles
planDesign.beamNames = beamNames
planDesign.calibration = ctCalibration
planDesign.xBeamletSpacing_mm = 5
planDesign.yBeamletSpacing_mm = 5
planDesign.targetMargin = 5.0
planDesign.defineTargetMaskAndPrescription(target = roi, targetPrescription = 20.)
plan = planDesign.buildPlan()
Load / Generate scenarios
scenario_folder = os.path.join(output_path, "RobustnessTest")
if os.path.isdir(scenario_folder):
scenarios = RobustnessEvalPhoton()
scenarios.selectionStrategy = RobustnessEvalPhoton.Strategies.DEFAULT
scenarios.setupSystematicError = plan.planDesign.robustnessEval.setupSystematicError
scenarios.setupRandomError = plan.planDesign.robustnessEval.setupRandomError
scenarios.load(scenario_folder)
else:
# Robust config for scenario dose computation
plan.planDesign.robustnessEval = RobustnessEvalPhoton()
plan.planDesign.robustnessEval.setupSystematicError = [1.6, 1.6, 1.6] #sigma (mm)
plan.planDesign.robustnessEval.setupRandomError = [1.4, 1.4, 1.4] #sigma (mm)
# Strategy selection
plan.planDesign.robustnessEval.selectionStrategy = plan.planDesign.robustnessEval.Strategies.REDUCED_SET
# plan.planDesign.robustnessEval.selectionStrategy = plan.planDesign.robustnessEval.Strategies.ALL
# plan.planDesign.robustnessEval.selectionStrategy = plan.planDesign.robustnessEval.Strategies.RANDOM
# plan.planDesign.robustnessEval.NumScenarios = 50
plan.planDesign.robustnessEval.doseDistributionType = "Nominal"
plan.patient = None
# run MCsquare simulation
scenarios = ccc.computeRobustScenario(ct, plan, roi = [roi], robustMode = "Shift") # 'Simulation' for total recomputation
output_folder = os.path.join(output_path, "RobustnessTest")
scenarios.save(output_folder)
Simplifying plan...
There are 2 beams per batch file
Robustness analysis
scenarios.recomputeDVH([roi])
scenarios.analyzeErrorSpace(ct, "D95", roi, plan.planDesign.objectives.targetPrescription)
scenarios.printInfo()
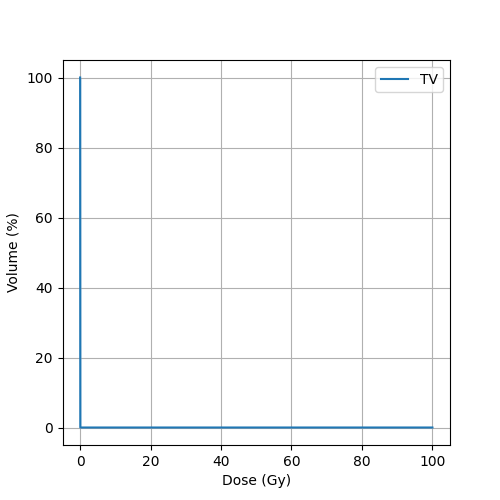
Display DVH + DVH-bands
fig, ax = plt.subplots(1, 1, figsize=(5, 5))
for dvh_band in scenarios.dvhBands:
phigh = ax.plot(dvh_band._dose, dvh_band._volumeHigh, alpha=0)
plow = ax.plot(dvh_band._dose, dvh_band._volumeLow, alpha=0)
pNominal = ax.plot(dvh_band._nominalDVH._dose, dvh_band._nominalDVH._volume, label=dvh_band._roiName, color = 'C0')
pfill = ax.fill_between(dvh_band._dose, dvh_band._volumeHigh, dvh_band._volumeLow, alpha=0.2, color='C0')
ax.set_xlabel("Dose (Gy)")
ax.set_ylabel("Volume (%)")
plt.grid(True)
plt.legend()
plt.savefig(os.path.join(output_path, 'Evaluation_RobustOptimizationPhotons.png'))
plt.show()
Total running time of the script: (2 minutes 38.185 seconds)
