Note
Go to the end to download the full example code.
Midp#
author: OpenTPS team
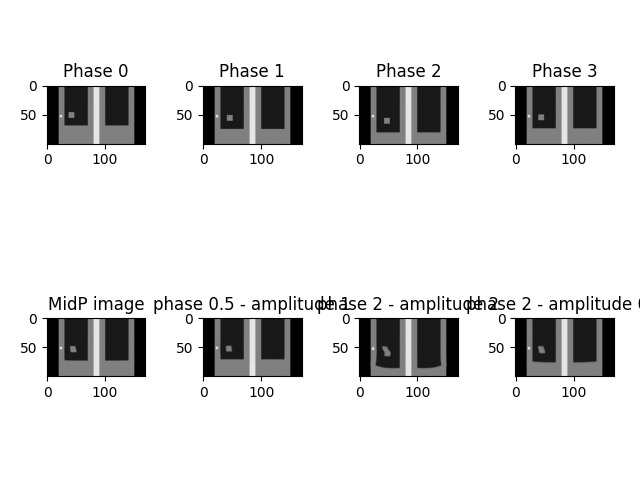
This example shows how to create a mid-position CT from a 4DCT and visualize it.
running time: ~ 6 minutes
Setting up the environment in google collab#
import sys
if "google.colab" in sys.modules:
from IPython import get_ipython
get_ipython().system('git clone https://gitlab.com/openmcsquare/opentps.git')
get_ipython().system('pip install ./opentps')
import opentps
imports
import numpy as np
import matplotlib.pyplot as plt
import time
import logging
import math
import os
import the needed opentps.core packages
from opentps.core.data.dynamicData._dynamic3DModel import Dynamic3DModel
from opentps.core.data.dynamicData._dynamic3DSequence import Dynamic3DSequence
from opentps.core.data.images import CTImage
from opentps.core.data.images import ROIMask
logger = logging.getLogger(__name__)
Output path#
output_path = os.path.join(os.getcwd(), 'Output', 'ExampleMidP')
if not os.path.exists(output_path):
os.makedirs(output_path)
logger.info('Files will be stored in {}'.format(output_path))
Synthetic 4DCT generation function#
def getPhasesPositions(numberOfPhases, minValue, maxValue):
angleList = np.linspace(0, 2 * math.pi, numberOfPhases + 1)[:-1]
cosList = np.cos(angleList)
diff = maxValue - minValue
posList = minValue + diff / 2 + cosList * diff / 2
return posList.astype(np.uint8)
def createSynthetic3DCT(diaphragmPos = 20, targetPos = [50, 100, 35], spacing=[1, 1, 2], returnTumorMask = False):
# GENERATE SYNTHETIC CT IMAGE
# background
im = np.full((170, 170, 100), -1000)
im[20:150, 70:130, :] = 0
# left lung
im[30:70, 80:120, diaphragmPos:] = -800
# right lung
im[100:140, 80:120, diaphragmPos:] = -800
# target
im[targetPos[0]-5:targetPos[0]+5, targetPos[1]-5:targetPos[1]+5, targetPos[2]-5:targetPos[2]+5] = 0
# vertebral column
im[80:90, 95:105, :] = 800
# rib
im[22:26, 90:110, 46:50] = 800
# couch
im[:, 130:135, :] = 100
ct = CTImage(imageArray=im, name='fixed', origin=[0, 0, 0], spacing=spacing)
if returnTumorMask:
mask = np.full((170, 170, 100), 0)
mask[targetPos[0]-5:targetPos[0]+5, targetPos[1]-5:targetPos[1]+5, targetPos[2]-5:targetPos[2]+5] = 1
roi = ROIMask(imageArray=mask, origin=[0, 0, 0], spacing=spacing)
return ct, roi
else:
return ct
def createSynthetic4DCT(numberOfPhases=4, spacing=[1, 1, 2], returnTumorMasks=False, motionNoise=True):
# GENERATE SYNTHETIC 4D INPUT SEQUENCE
CT4D = Dynamic3DSequence()
## For the diaphragm position
diaphMotionAmp = 12
diaphMinPos = 20
diaphPosList = getPhasesPositions(numberOfPhases, diaphMinPos, diaphMinPos+diaphMotionAmp)
if motionNoise:
diaphNoise = [[3, 1],
[6, -1],
[9, -1],
[12, 1],
[15, 1]]
else:
diaphNoise = [[3, 0],
[6, 0],
[9, 0],
[12, 0],
[15, 0]]
for elemIdx in range(len(diaphNoise)):
if diaphNoise[elemIdx][0] <= numberOfPhases - 1:
diaphPosList[diaphNoise[elemIdx][0]] += diaphNoise[elemIdx][1]
## For the target z position
zMotionAmp = int(np.round(diaphMotionAmp * 0.8))
zMinPos = 40
zPosList = getPhasesPositions(numberOfPhases, zMinPos, zMinPos+zMotionAmp)
if motionNoise:
zNoise = [[3, 1],
[6, -1],
[9, -1],
[12, 1],
[15, 1]]
else:
zNoise = [[3, 0],
[6, 0],
[9, 0],
[12, 0],
[15, 0]]
for elemIdx in range(len(zNoise)):
if zNoise[elemIdx][0] <= numberOfPhases - 1:
zPosList[zNoise[elemIdx][0]] += zNoise[elemIdx][1]
## For the target x position
xMotionAmp = 6
xMinPos = 42
xPosList = getPhasesPositions(numberOfPhases, xMinPos, xMinPos+xMotionAmp)
if motionNoise:
xNoise = [[3, 1],
[6, -1],
[9, -1],
[12, 1],
[15, 1]]
else:
xNoise = [[3, 0],
[6, 0],
[9, 0],
[12, 0],
[15, 0]]
for elemIdx in range(len(xNoise)):
if xNoise[elemIdx][0] <= numberOfPhases-1:
xPosList[xNoise[elemIdx][0]] += xNoise[elemIdx][1]
xPosList = np.roll(xPosList, 2)
# print('xPosList', xPosList)
phaseList = []
if returnTumorMasks:
maskList = []
for phaseIndex in range(numberOfPhases):
phase, mask = createSynthetic3DCT(targetPos=[xPosList[phaseIndex], 95, zPosList[phaseIndex]], diaphragmPos=diaphPosList[phaseIndex], spacing=spacing, returnTumorMask=returnTumorMasks)
phaseList.append(phase)
maskList.append(mask)
else:
for phaseIndex in range(numberOfPhases):
phase = createSynthetic3DCT(targetPos=[xPosList[phaseIndex], 95, zPosList[phaseIndex]], diaphragmPos=diaphPosList[phaseIndex], spacing=spacing)
phaseList.append(phase)
CT4D.dyn3DImageList = phaseList
if returnTumorMasks:
return CT4D, maskList
else:
return CT4D
Generate synthetic 4DCT And MidP#
# GENERATE SYNTHETIC 4D INPUT SEQUENCE
CT4D = createSynthetic4DCT()
# GENERATE MIDP
Model4D = Dynamic3DModel()
startTime = time.time()
Model4D.computeMidPositionImage(CT4D, 0, tryGPU=True)
stopTime = time.time()
print('midP computed in ', np.round(stopTime - startTime, 2), 'seconds')
# GENERATE ADDITIONAL PHASES
im1 = Model4D.generate3DImage(0.5/4, amplitude=1, tryGPU=False)
im2 = Model4D.generate3DImage(2/4, amplitude=2.0, tryGPU=False)
im3 = Model4D.generate3DImage(2/4, amplitude=0.5, tryGPU=False)
Dynamic 3D Sequence Created with 0 images
midP computed in 15.81 seconds
Display results#
fig, ax = plt.subplots(2, 4)
fig.tight_layout()
y_slice = 95
ax[0,0].imshow(CT4D.dyn3DImageList[0].imageArray[:, y_slice, :].T[::-1, ::1], cmap='gray', origin='upper', vmin=-1000, vmax=1000)
ax[0,0].title.set_text('Phase 0')
ax[0,1].imshow(CT4D.dyn3DImageList[1].imageArray[:, y_slice, :].T[::-1, ::1], cmap='gray', origin='upper', vmin=-1000, vmax=1000)
ax[0,1].title.set_text('Phase 1')
ax[0,2].imshow(CT4D.dyn3DImageList[2].imageArray[:, y_slice, :].T[::-1, ::1], cmap='gray', origin='upper', vmin=-1000, vmax=1000)
ax[0,2].title.set_text('Phase 2')
ax[0,3].imshow(CT4D.dyn3DImageList[3].imageArray[:, y_slice, :].T[::-1, ::1], cmap='gray', origin='upper', vmin=-1000, vmax=1000)
ax[0,3].title.set_text('Phase 3')
ax[1,0].imshow(Model4D.midp.imageArray[:, y_slice, :].T[::-1, ::1], cmap='gray', origin='upper', vmin=-1000, vmax=1000)
ax[1,0].title.set_text('MidP image')
ax[1,1].imshow(im1.imageArray[:, y_slice, :].T[::-1, ::1], cmap='gray', origin='upper', vmin=-1000, vmax=1000)
ax[1,1].title.set_text('phase 0.5 - amplitude 1')
ax[1,2].imshow(im2.imageArray[:, y_slice, :].T[::-1, ::1], cmap='gray', origin='upper', vmin=-1000, vmax=1000)
ax[1,2].title.set_text('phase 2 - amplitude 2')
ax[1,3].imshow(im3.imageArray[:, y_slice, :].T[::-1, ::1], cmap='gray', origin='upper', vmin=-1000, vmax=1000)
ax[1,3].title.set_text('phase 2 - amplitude 0.5')
plt.savefig(os.path.join(output_path, 'ExampleMidp.png'))
print('MidP example completed')
plt.show()
MidP example completed
Total running time of the script: (0 minutes 17.530 seconds)
