Note
Go to the end to download the full example code.
Deformable Breathing Data Augmentation#
author: OpenTPS team
This example shows how to create a synthetic 4DCT, generate a mid-position CT, and create a dynamic sequence from breathing signals and the mid-position CT. The example also demonstrates how to visualize the generated dynamic sequence.
running time: ~ 7 minutes
Setting up the environment in google collab#
import sys
if "google.colab" in sys.modules:
from IPython import get_ipython
get_ipython().system('git clone https://gitlab.com/openmcsquare/opentps.git')
get_ipython().system('pip install ./opentps')
import opentps
imports
import os
import sys
currentWorkingDir = os.getcwd()
sys.path.append(currentWorkingDir)
import numpy as np
import math
import logging
import matplotlib.pyplot as plt
import the needed opentps.core packages
from matplotlib.animation import FuncAnimation
from opentps.core.data.dynamicData._breathingSignals import SyntheticBreathingSignal
from opentps.core.data.dynamicData._dynamic3DModel import Dynamic3DModel
from opentps.core.data.dynamicData._dynamic3DSequence import Dynamic3DSequence
from opentps.core.processing.deformableDataAugmentationToolBox.generateDynamicSequencesFromModel import generateDynSeqFromBreathingSignalsAndModel
from opentps.core.processing.imageProcessing.imageTransform3D import getVoxelIndexFromPosition
from opentps.core.processing.imageProcessing.resampler3D import resample
from opentps.core.data.images import CTImage
from opentps.core.data.images import ROIMask
logger = logging.getLogger(__name__)
Output path#
output_path = os.path.join(os.getcwd(), 'Output', 'ExampleDeformableBreathingDataAugmentation')
if not os.path.exists(output_path):
os.makedirs(output_path)
logger.info('Files will be stored in {}'.format(output_path))
Synthetic 4DCT generation function#
def getPhasesPositions(numberOfPhases, minValue, maxValue):
angleList = np.linspace(0, 2 * math.pi, numberOfPhases + 1)[:-1]
cosList = np.cos(angleList)
diff = maxValue - minValue
posList = minValue + diff / 2 + cosList * diff / 2
return posList.astype(np.uint8)
def createSynthetic3DCT(diaphragmPos = 20, targetPos = [50, 100, 35], spacing=[1, 1, 2], returnTumorMask = False):
# GENERATE SYNTHETIC CT IMAGE
# background
im = np.full((170, 170, 100), -1000)
im[20:150, 70:130, :] = 0
# left lung
im[30:70, 80:120, diaphragmPos:] = -800
# right lung
im[100:140, 80:120, diaphragmPos:] = -800
# target
im[targetPos[0]-5:targetPos[0]+5, targetPos[1]-5:targetPos[1]+5, targetPos[2]-5:targetPos[2]+5] = 0
# vertebral column
im[80:90, 95:105, :] = 800
# rib
im[22:26, 90:110, 46:50] = 800
# couch
im[:, 130:135, :] = 100
ct = CTImage(imageArray=im, name='fixed', origin=[0, 0, 0], spacing=spacing)
if returnTumorMask:
mask = np.full((170, 170, 100), 0)
mask[targetPos[0]-5:targetPos[0]+5, targetPos[1]-5:targetPos[1]+5, targetPos[2]-5:targetPos[2]+5] = 1
roi = ROIMask(imageArray=mask, origin=[0, 0, 0], spacing=spacing)
return ct, roi
else:
return ct
def createSynthetic4DCT(numberOfPhases=4, spacing=[1, 1, 2], returnTumorMasks=False, motionNoise=True):
# GENERATE SYNTHETIC 4D INPUT SEQUENCE
CT4D = Dynamic3DSequence()
## For the diaphragm position
diaphMotionAmp = 12
diaphMinPos = 20
diaphPosList = getPhasesPositions(numberOfPhases, diaphMinPos, diaphMinPos+diaphMotionAmp)
if motionNoise:
diaphNoise = [[3, 1],
[6, -1],
[9, -1],
[12, 1],
[15, 1]]
else:
diaphNoise = [[3, 0],
[6, 0],
[9, 0],
[12, 0],
[15, 0]]
for elemIdx in range(len(diaphNoise)):
if diaphNoise[elemIdx][0] <= numberOfPhases - 1:
diaphPosList[diaphNoise[elemIdx][0]] += diaphNoise[elemIdx][1]
## For the target z position
zMotionAmp = int(np.round(diaphMotionAmp * 0.8))
zMinPos = 40
zPosList = getPhasesPositions(numberOfPhases, zMinPos, zMinPos+zMotionAmp)
if motionNoise:
zNoise = [[3, 1],
[6, -1],
[9, -1],
[12, 1],
[15, 1]]
else:
zNoise = [[3, 0],
[6, 0],
[9, 0],
[12, 0],
[15, 0]]
for elemIdx in range(len(zNoise)):
if zNoise[elemIdx][0] <= numberOfPhases - 1:
zPosList[zNoise[elemIdx][0]] += zNoise[elemIdx][1]
## For the target x position
xMotionAmp = 6
xMinPos = 42
xPosList = getPhasesPositions(numberOfPhases, xMinPos, xMinPos+xMotionAmp)
if motionNoise:
xNoise = [[3, 1],
[6, -1],
[9, -1],
[12, 1],
[15, 1]]
else:
xNoise = [[3, 0],
[6, 0],
[9, 0],
[12, 0],
[15, 0]]
for elemIdx in range(len(xNoise)):
if xNoise[elemIdx][0] <= numberOfPhases-1:
xPosList[xNoise[elemIdx][0]] += xNoise[elemIdx][1]
xPosList = np.roll(xPosList, 2)
# print('xPosList', xPosList)
phaseList = []
if returnTumorMasks:
maskList = []
for phaseIndex in range(numberOfPhases):
phase, mask = createSynthetic3DCT(targetPos=[xPosList[phaseIndex], 95, zPosList[phaseIndex]], diaphragmPos=diaphPosList[phaseIndex], spacing=spacing, returnTumorMask=returnTumorMasks)
phaseList.append(phase)
maskList.append(mask)
else:
for phaseIndex in range(numberOfPhases):
phase = createSynthetic3DCT(targetPos=[xPosList[phaseIndex], 95, zPosList[phaseIndex]], diaphragmPos=diaphPosList[phaseIndex], spacing=spacing)
phaseList.append(phase)
CT4D.dyn3DImageList = phaseList
if returnTumorMasks:
return CT4D, maskList
else:
return CT4D
Generate synthetic 4DCT#
CT4D = createSynthetic4DCT()
plt.figure()
fig = plt.gcf()
def updateAnim(imageIndex):
fig.clear()
plt.imshow(np.rot90(CT4D.dyn3DImageList[imageIndex].imageArray[:, 95, :]))
anim = FuncAnimation(fig, updateAnim, frames=len(CT4D.dyn3DImageList), interval=300)
anim.save(os.path.join(output_path, 'anim.gif'))
plt.show()
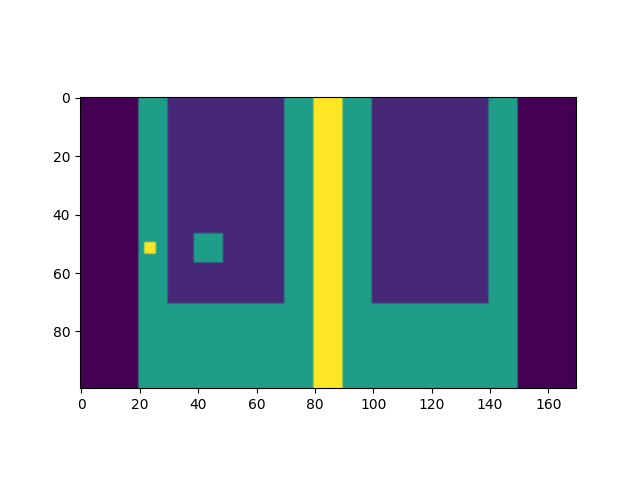
Dynamic 3D Sequence Created with 0 images
Generate MidP#
dynMod = Dynamic3DModel()
dynMod.computeMidPositionImage(CT4D, 0, tryGPU=True)
print(dynMod.midp.origin, dynMod.midp.spacing, dynMod.midp.gridSize)
print('Resample model image')
dynMod = resample(dynMod, gridSize=(80, 50, 50))
print('after resampling', dynMod.midp.origin, dynMod.midp.spacing, dynMod.midp.gridSize)
# option 3
for field in dynMod.deformationList:
print('Resample model field')
field.resample(spacing=dynMod.midp.spacing, gridSize=dynMod.midp.gridSize, origin=dynMod.midp.origin)
print('after resampling', field.origin, field.spacing, field.gridSize)
simulationTime = 10
amplitude = 10
newSignal = SyntheticBreathingSignal(amplitude=amplitude,
breathingPeriod=4,
meanNoise=0,
varianceNoise=0,
samplingPeriod=0.2,
simulationTime=simulationTime,
coeffMin=0,
coeffMax=0,
meanEvent=0/30,
meanEventApnea=0)
newSignal.generate1DBreathingSignal()
linearIncrease = np.linspace(0.8, 10, newSignal.breathingSignal.shape[0])
newSignal.breathingSignal = newSignal.breathingSignal * linearIncrease
newSignal2 = SyntheticBreathingSignal()
newSignal2.breathingSignal = -newSignal.breathingSignal
signalList = [newSignal.breathingSignal, newSignal2.breathingSignal]
pointRLung = np.array([50, 100, 50])
pointLLung = np.array([120, 100, 50])
## get points in voxels --> for the plot, not necessary for the process example
pointRLungInVoxel = getVoxelIndexFromPosition(pointRLung, dynMod.midp)
pointLLungInVoxel = getVoxelIndexFromPosition(pointLLung, dynMod.midp)
pointList = [pointRLung, pointLLung]
pointVoxelList = [pointRLungInVoxel, pointLLungInVoxel]
## to show signals and ROIs
prop_cycle = plt.rcParams['axes.prop_cycle']
colors = prop_cycle.by_key()['color']
plt.figure(figsize=(12, 6))
signalAx = plt.subplot(2, 1, 2)
for pointIndex, point in enumerate(pointList):
ax = plt.subplot(2, 2 * len(pointList), 2 * pointIndex + 1)
ax.set_title('Slice Y:' + str(pointVoxelList[pointIndex][1]))
ax.imshow(np.rot90(dynMod.midp.imageArray[:, pointVoxelList[pointIndex][1], :]))
ax.scatter([pointVoxelList[pointIndex][0]], [dynMod.midp.imageArray.shape[2] - pointVoxelList[pointIndex][2]], c=colors[pointIndex], marker="x", s=100)
ax2 = plt.subplot(2, 2 * len(pointList), 2 * pointIndex + 2)
ax2.set_title('Slice Z:' + str(pointVoxelList[pointIndex][2]))
ax2.imshow(np.rot90(dynMod.midp.imageArray[:, :, pointVoxelList[pointIndex][2]], 3))
ax2.scatter([pointVoxelList[pointIndex][0]], [pointVoxelList[pointIndex][1]], c=colors[pointIndex], marker="x", s=100)
signalAx.plot(newSignal.timestamps / 1000, signalList[pointIndex], c=colors[pointIndex])
signalAx.set_xlabel('Time (s)')
signalAx.set_ylabel('Deformation amplitude in Z direction (mm)')
plt.show()
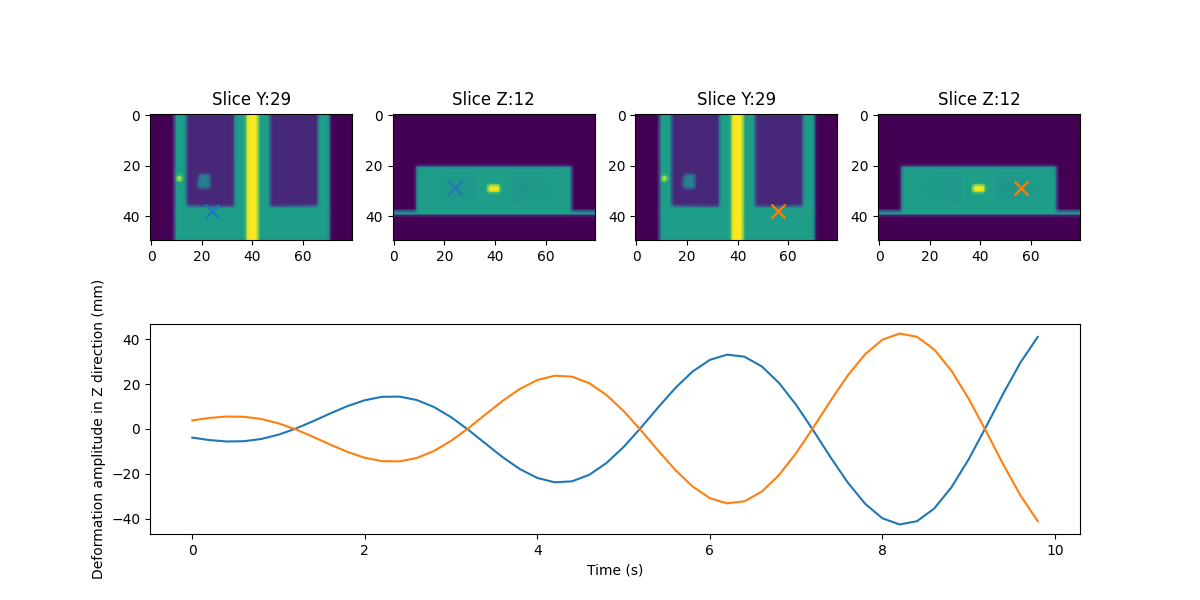
[0 0 0] [1 1 2] [170 170 100]
Resample model image
after resampling [0 0 0] [2.125 3.4 4. ] [80 50 50]
Resample model field
after resampling [0 0 0] [2.125 3.4 4. ] [80 50 50]
Resample model field
after resampling [0 0 0] [2.125 3.4 4. ] [80 50 50]
Resample model field
after resampling [0 0 0] [2.125 3.4 4. ] [80 50 50]
Resample model field
after resampling [0 0 0] [2.125 3.4 4. ] [80 50 50]
dynSeq = generateDynSeqFromBreathingSignalsAndModel(dynMod, signalList, pointList, dimensionUsed='Z', outputType=np.int16)
dynSeq.breathingPeriod = newSignal.breathingPeriod
dynSeq.timingsList = newSignal.timestamps
print('/'*80, '\n', '/'*80)
plt.figure()
fig = plt.gcf()
def updateAnim(imageIndex):
fig.clear()
plt.imshow(np.rot90(dynSeq.dyn3DImageList[imageIndex].imageArray[:, 29, :]))
anim = FuncAnimation(fig, updateAnim, frames=len(dynSeq.dyn3DImageList), interval=300)
anim.save(os.path.join(output_path, 'anim3.gif'))
plt.show()
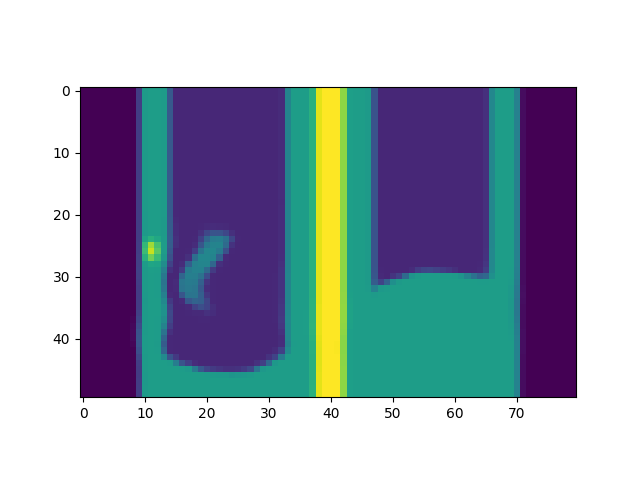
Signal indexes used [0, 50]
Compute displacement field from velocity field for field 0
Compute displacement field from velocity field for field 1
Compute displacement field from velocity field for field 2
Compute displacement field from velocity field for field 3
Dynamic 3D Sequence Created with 0 images
Deform image 0
Deform image 1
Deform image 2
Deform image 3
Deform image 4
Deform image 5
Deform image 6
Deform image 7
Deform image 8
Deform image 9
Deform image 10
Deform image 11
Deform image 12
Deform image 13
Deform image 14
Deform image 15
Deform image 16
Deform image 17
Deform image 18
Deform image 19
Deform image 20
Deform image 21
Deform image 22
Deform image 23
Deform image 24
Deform image 25
Deform image 26
Deform image 27
Deform image 28
Deform image 29
Deform image 30
Deform image 31
Deform image 32
Deform image 33
Deform image 34
Deform image 35
Deform image 36
Deform image 37
Deform image 38
Deform image 39
Deform image 40
Deform image 41
Deform image 42
Deform image 43
Deform image 44
Deform image 45
Deform image 46
Deform image 47
Deform image 48
Deform image 49
////////////////////////////////////////////////////////////////////////////////
////////////////////////////////////////////////////////////////////////////////
Total running time of the script: (0 minutes 29.368 seconds)
