Note
Go to the end to download the full example code.
Use of Range Shifter for Proton Plan#
author: OpenTPS team
In this example, we will show how to create a plan from scratch and use range shifters.
running time: ~ 10 minutes
Setting up the environment in google collab#
import sys
if "google.colab" in sys.modules:
from IPython import get_ipython
get_ipython().system('git clone https://gitlab.com/openmcsquare/opentps.git')
get_ipython().system('pip install ./opentps')
import opentps
imports
import os
import logging
import sys
from matplotlib import pyplot as plt
import the needed opentps.core packages
from opentps.core.processing.imageProcessing.resampler3D import resampleImage3DOnImage3D
sys.path.append('..')
import numpy as np
from pathlib import Path
from opentps.core.data.plan._planProtonBeam import PlanProtonBeam
from opentps.core.data.plan._planProtonLayer import PlanProtonLayer
from opentps.core.data.plan._protonPlan import ProtonPlan
from opentps.core.data.plan._rtPlan import RTPlan
from opentps.core.io.scannerReader import readScanner
from opentps.core.io.serializedObjectIO import loadRTPlan, saveRTPlan
from opentps.core.io import mcsquareIO
from opentps.core.data._dvh import DVH
from opentps.core.processing.doseCalculation.doseCalculationConfig import DoseCalculationConfig
from opentps.core.io.dataLoader import readData
from opentps.core.io.mhdIO import exportImageMHD
from opentps.core.processing.doseCalculation.protons.mcsquareDoseCalculator import MCsquareDoseCalculator
from opentps.core.data.images import CTImage, DoseImage
from opentps.core.data.images import ROIMask
from opentps.core.io.dicomIO import writeDicomCT, writeRTPlan, writeRTDose, readDicomDose, writeRTStruct
from opentps.core.io.mcsquareIO import RangeShifter
from opentps.core.data.CTCalibrations.MCsquareCalibration._mcsquareMolecule import MCsquareMolecule
from opentps.core.data._rtStruct import RTStruct
from opentps.core.data import Patient
logger = logging.getLogger(__name__)
Output path#
output_path = os.path.join(os.getcwd(), 'Exemple_RangeShifter')
if not os.path.exists(output_path):
os.makedirs(output_path)
print(f"Directory '{output_path}' created.")
else:
print(f"Directory '{output_path}' already exists.")
logger.info('Files will be stored in {}'.format(output_path))
Directory '/home/runner/work/examples/examples/examples/DoseComputation/Exemple_RangeShifter' created.
Choosing default scanner
doseCalculator = MCsquareDoseCalculator()
doseCalculator.ctCalibration = readScanner(DoseCalculationConfig().scannerFolder)
# Or a specific one if you do have
# MCSquarePath = os.path.join(openTPS_path, 'core', 'processing', 'doseCalculation', 'MCsquare')
# scannerPath = os.path.join(MCSquarePath, 'Scanners', 'UCL_Toshiba')
# doseCalculator.ctCalibration = MCsquareCTCalibration(fromFiles=(os.path.join(scannerPath, 'HU_Density_Conversion.txt'),
# os.path.join(scannerPath, 'HU_Material_Conversion.txt'),
# os.path.join(MCSquarePath, 'Materials')))
Path of your BDL if you do have one Otherwise the default bdl ‘BDL_default_DN_RangeShifter’ is used
bdl_path = "Path/to/your/BDL"
if 'bdl_path' in locals() and os.path.isfile(bdl_path):
DoseCalculationConfig().bdlFile = bdl_path
# chossing default BDL
doseCalculator.beamModel = mcsquareIO.readBDL(DoseCalculationConfig().bdlFile)
Get different range shifters#
From bdl file : Be sure the material you use is in core/processing/doseCalculation/protons/MCsquare/Materials and you have the good MCsquare material ID in the bdl If you want to add a new material, you can add the folder with the necessary material properties in MCsquare/Materials The MCsquare material ID (RS_material) of the new material to add in the bdl will be print in the terminal and is in MCsquare/Materials/list.dat
rs_1 = doseCalculator.beamModel.rangeShifters[0]
# From scratch :
rs_2 = RangeShifter(material='Lexan', density=1.217, WET=40.3)
rs_2.ID = 'RS_Lexan_66'
rs_2.type = 'binary'
print('Range shifter 1:', rs_1)
print('Range shifter 2:', rs_2)
Range shifter 1: RS_ID = RS_Block
RS_type = binary
RS_density = 1.2
RS_WET = 74.1
Range shifter 2: RS_ID = RS_Lexan_66
RS_type = binary
RS_density = 1.217
RS_WET = 40.3
Configure dose calculation#
doseCalculator.nbPrimaries = 1e7 # number of primary particles, 1e4 is enough for a quick test, otherwise 1e7 is recommended (It can take several minutes to compute).
patient = Patient()
patient.name = 'TestPatient'
Define CT and Target#
ctSize = 200
ct = CTImage()
ct.name = 'TestPhantom'
ct.patient = patient
target = ROIMask()
target.name = 'TV'
target.spacing = ct.spacing
target.color = (255, 0, 0) # red
targetArray = np.zeros((ctSize, ctSize, ctSize)).astype(bool)
radius = 20
x0, y0, z0 = (100, 100, 100)
x, y, z = np.mgrid[0:ctSize:1, 0:ctSize:1, 0:ctSize:1]
r = np.sqrt((x - x0) ** 2 + (y - y0) ** 2 + (z - z0) ** 2)
targetArray[r < radius] = True
target.imageArray = targetArray
huAir = -1024.
huWater = doseCalculator.ctCalibration.convertRSP2HU(1.)
ctArray = huAir * np.ones((ctSize, ctSize, ctSize))
ctArray[1:ctSize - 1, 1:ctSize - 1, 1:ctSize - 1] = huWater
ctArray[targetArray >= 0.5] = 50
ct.imageArray = ctArray
body = ROIMask()
body.name = 'Body'
body.spacing = ct.spacing
body.color = (0, 0, 255)
bodyArray = np.zeros((ctSize, ctSize, ctSize)).astype(bool)
bodyArray[1:ctSize - 1, 1:ctSize - 1, 1:ctSize - 1] = True
body.imageArray = bodyArray
Create plan from scratch#
plan = ProtonPlan()
plan.appendBeam(PlanProtonBeam())
plan.appendBeam(PlanProtonBeam())
plan.appendBeam(PlanProtonBeam())
plan.beams[0].gantryAngle = 0.
plan.beams[1].gantryAngle = 0.
plan.beams[2].gantryAngle = 90.
plan.beams[0].appendLayer(PlanProtonLayer(120)) # Nominal energy of the layer
plan.beams[1].appendLayer(PlanProtonLayer(120))
plan.beams[2].appendLayer(PlanProtonLayer(120))
plan[0].layers[0].appendSpot([50, 60], [100, 100], [300, 300]) # Two spots to the target from beam 0 (0. gantryAngle)
plan[1].layers[0].appendSpot([90, 100], [100, 100], [300, 300]) # Two spots to the target from beam 1 (0. gantryAngle)
plan[2].layers[0].appendSpot([100, 110], [100, 110], [300, 300]) # Two spots placed outside the target from beam 2 (90. gantryAngle)
Use 2 different range shifters for the two beams
plan.rangeShifter = [rs_1, rs_2]
plan.beams[0].rangeShifter = [rs_1]
plan.beams[1].rangeShifter = None
plan.beams[2].rangeShifter = [rs_2]
Range Shifter beam 0 parameters
plan[0].layers[0].rangeShifterSettings.isocenterToRangeShifterDistance = 0 # [mm]
plan[0].layers[0].rangeShifterSettings.rangeShifterSetting = 'IN'
plan[0].layers[0].rangeShifterSettings.rangeShifterWaterEquivalentThickness = None # [mm] None means get thickness from BDL
Range Shifter beam 2 parameters
plan[2].layers[0].rangeShifterSettings.isocenterToRangeShifterDistance = 200 # [mm]
plan[2].layers[0].rangeShifterSettings.rangeShifterSetting = 'IN'
plan[2].layers[0].rangeShifterSettings.rangeShifterWaterEquivalentThickness = 15 # [mm] None means get thickness from BDL
Save plan in OpenTPS format (serialized)
saveRTPlan(plan, os.path.join(output_path, 'dummy_plan.tps'))
Load plan in OpenTPS format (serialized)
plan2 = loadRTPlan(os.path.join(output_path, 'dummy_plan.tps'))
print(plan2[0].layers[0].spotWeights)
print(plan[0].layers[0].spotWeights) # plan2 is the same as plan
[300. 300.]
[300. 300.]
Save plan in Dicom format
dicomPath = os.path.join(output_path)
writeRTPlan(plan, dicomPath)
writeDicomCT(ct, os.path.join(dicomPath, 'CT'))
print('Dicom files saved in', dicomPath)
# For contour, they must be RTStruct
contour = target.getROIContour()
struct = RTStruct()
struct.appendContour(contour)
writeRTStruct(struct, os.path.join(dicomPath, 'CT'))
# load plan in Dicom format
dataList = readData(dicomPath, maxDepth=2)
ctDicom = [d for d in dataList if isinstance(d, CTImage)][0]
planDicom = [d for d in dataList if isinstance(d, RTPlan)][0]
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/pydicom/valuerep.py:440: UserWarning: Invalid value for VR CS: 'RT Ion Plan IOD'. Please see <https://dicom.nema.org/medical/dicom/current/output/html/part05.html#table_6.2-1> for allowed values for each VR.
warn_and_log(msg)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/pydicom/dataset.py:2710: UserWarning: Camel case attribute 'PrivateCreator' used which is not in the element keyword data dictionary
warn_and_log(msg)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/pydicom/valuerep.py:440: UserWarning: A value of type 'int64' cannot be assigned to a tag with VR US.
warn_and_log(msg)
Dicom files saved in /home/runner/work/examples/examples/examples/DoseComputation/Exemple_RangeShifter
Generic example: box of water with spherical target#
# Load CT & contours
# ct = [d for d in dataList if isinstance(d, CTImage)][0]
# struct = [d for d in dataList if isinstance(d, RTStruct)][0]
# target = struct.getContourByName('TV')
# body = struct.getContourByName('Body')
# Compute the dose
doseImage = doseCalculator.computeDose(ct, plan) # You can choose the plan you want to use, results will be the same
# doseImage = importImageMHD(output_path) # If you want to import a dose image from a MHD file
# doseImageDicom = [d for d in dataList if isinstance(d, DoseImage)][0] # If you want to import a dose image from a Dicom file
# Export dose (MHD)
# exportImageMHD(os.path.join(output_path,'DoseImage'), doseImage)
# Export dose (Dicom)
writeRTDose(doseImage, dicomPath)
# doseImageDicom = readDicomDose(r"pathToDicomDose") # If you want to import a dose image from a Dicom file
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/pydicom/dataset.py:2710: UserWarning: Camel case attribute 'SoftwareVersion' used which is not in the element keyword data dictionary
warn_and_log(msg)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/pydicom/dataset.py:2710: UserWarning: Camel case attribute 'OperatorName' used which is not in the element keyword data dictionary
warn_and_log(msg)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/pydicom/dataset.py:2710: UserWarning: Camel case attribute 'Width' used which is not in the element keyword data dictionary
warn_and_log(msg)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/pydicom/valuerep.py:440: UserWarning: A value of type 'int64' cannot be assigned to a tag with VR US.
warn_and_log(msg)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/pydicom/dataset.py:2710: UserWarning: Camel case attribute 'Height' used which is not in the element keyword data dictionary
warn_and_log(msg)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/pydicom/dataset.py:2710: UserWarning: Camel case attribute 'ColorType' used which is not in the element keyword data dictionary
warn_and_log(msg)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/pydicom/dataset.py:2710: UserWarning: Camel case attribute 'BitDepth' used which is not in the element keyword data dictionary
warn_and_log(msg)
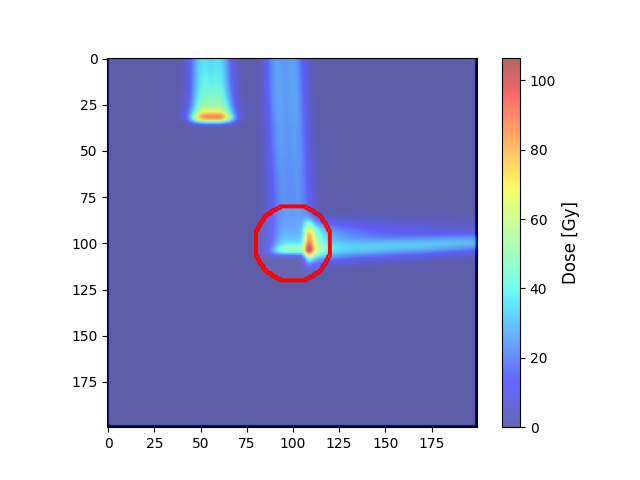
Plot dose#
target = resampleImage3DOnImage3D(target, ct)
COM_coord = target.centerOfMass
COM_index = target.getVoxelIndexFromPosition(COM_coord)
Z_coord = COM_index[2]
img_ct = ct.imageArray[:, :, Z_coord].transpose(1, 0)
contourTargetMask = target.getBinaryContourMask()
img_mask = contourTargetMask.imageArray[:, :, Z_coord].transpose(1, 0)
img_dose = resampleImage3DOnImage3D(doseImage, ct)
img_dose = img_dose.imageArray[:, :, Z_coord].transpose(1, 0)
Display dose#
plt.imshow(img_ct, cmap='gray')
plt.contour(img_mask, colors='red') # PTV
dose = plt.imshow(img_dose, cmap='jet', alpha=.6)
colorbar = plt.colorbar(dose)
colorbar.set_label('Dose [Gy]', fontsize=12)
plt.show()
plt.savefig(os.path.join(output_path, 'Dose_protonWithRangeShifters.png'))
Total running time of the script: (2 minutes 23.247 seconds)
