Note
Go to the end to download the full example code.
Simple IMPT proton plan optimization#
author: OpenTPS team
In this example, we will create and optimize a simple Protons plan.
running time: ~ 12 minutes
Setting up the environment in google collab#
import sys
if "google.colab" in sys.modules:
from IPython import get_ipython
get_ipython().system('git clone https://gitlab.com/openmcsquare/opentps.git')
get_ipython().system('pip install ./opentps')
import opentps
imports
import math
import os
import sys
import numpy as np
from matplotlib import pyplot as plt
import the needed opentps.core packages
import opentps.core.processing.planOptimization.objectives.dosimetricObjectives as doseObj
from opentps.core.data.images import CTImage
from opentps.core.data.images import ROIMask
from opentps.core.data.plan import ObjectivesList
from opentps.core.data.plan import ProtonPlanDesign
from opentps.core.data import DVH
from opentps.core.data import Patient
from opentps.core.io import mcsquareIO
from opentps.core.io.scannerReader import readScanner
from opentps.core.io.serializedObjectIO import saveRTPlan, loadRTPlan
from opentps.core.processing.doseCalculation.doseCalculationConfig import DoseCalculationConfig
from opentps.core.processing.doseCalculation.protons.mcsquareDoseCalculator import MCsquareDoseCalculator
from opentps.core.processing.imageProcessing.resampler3D import resampleImage3DOnImage3D, resampleImage3D
from opentps.core.processing.planOptimization.planOptimization import IntensityModulationOptimizer
CT calibration and BDL#
ctCalibration = readScanner(DoseCalculationConfig().scannerFolder)
bdl = mcsquareIO.readBDL(DoseCalculationConfig().bdlFile)
Create synthetic CT and ROI#
patient = Patient()
patient.name = 'Patient'
ctSize = 150
ct = CTImage()
ct.name = 'CT'
ct.patient = patient
huAir = -1024.
huWater = ctCalibration.convertRSP2HU(1.)
data = huAir * np.ones((ctSize, ctSize, ctSize))
data[:, 50:, :] = huWater
ct.imageArray = data
roi = ROIMask()
roi.patient = patient
roi.name = 'TV'
roi.color = (255, 0, 0) # red
data = np.zeros((ctSize, ctSize, ctSize)).astype(bool)
data[100:120, 100:120, 100:120] = True
roi.imageArray = data
# roi2 = ROIMask()
# roi2.patient = patient
# roi2.name = 'TV2'
# roi2.color = (0, 0, 255) # blue
# data = np.zeros((ctSize, ctSize, ctSize)).astype(bool)
# data[100:120, 70:100, 70:120] = True
# roi2.imageArray = data
body = roi.copy()
body.name = 'Body'
body.dilateMask(20)
body.imageArray = np.logical_xor(body.imageArray, roi.imageArray).astype(bool)
Configure dose engine#
mc2 = MCsquareDoseCalculator()
mc2.beamModel = bdl
mc2.nbPrimaries = 5e4
mc2.ctCalibration = ctCalibration
mc2._independentScoringGrid = True
scoringSpacing = [2,2,2]
mc2._scoringVoxelSpacing = scoringSpacing
Design plan#
beamNames = ["Beam1"]
gantryAngles = [0.]
couchAngles = [0.]
planInit = ProtonPlanDesign()
planInit.ct = ct
planInit.gantryAngles = gantryAngles
planInit.beamNames = beamNames
planInit.couchAngles = couchAngles
planInit.calibration = ctCalibration
planInit.spotSpacing = 5.0
planInit.layerSpacing = 5.0
planInit.targetMargin = 2.0
planInit.setScoringParameters(scoringSpacing=[2, 2, 2], adapt_gridSize_to_new_spacing=True)
# needs to be called after scoringGrid settings but prior to spot placement
planInit.defineTargetMaskAndPrescription(target = roi, targetPrescription = 20.)
plan = planInit.buildPlan() # Spot placement
plan.PlanName = "NewPlan"
beamlets = mc2.computeBeamlets(ct, plan, roi=[roi,body])
plan.planDesign.beamlets = beamlets
# doseImageRef = beamlets.toDoseImage()
objectives#
plan.planDesign.objectives.addObjective(doseObj.DMax(body,5, weight=1.0))
plan.planDesign.objectives.addObjective(doseObj.DMax(roi, 21, weight=10.0))
plan.planDesign.objectives.addObjective(doseObj.DMin(roi, 20, weight=20.0))
# Other examples of objectives
# plan.planDesign.objectives.addObjective(doseObj.DUniform(roi, 20, weight=10))
#
# plan.planDesign.objectives.addObjective(doseObj.DMaxMean(roi,21,weight=5))
# plan.planDesign.objectives.addObjective(doseObj.DMinMean(roi,20,weight=5))
# plan.planDesign.objectives.addObjective(doseObj.DFallOff(roi,oar,10,5,15,weight=5))
#
# plan.planDesign.objectives.addObjective(doseObj.DVHMax(roi, 21,0.05, weight=5))
# plan.planDesign.objectives.addObjective(doseObj.DVHMin(roi, 18,0.90, weight=5))
#
# plan.planDesign.objectives.addObjective(doseObj.EUDMin(roi, 19, 1, weight=50))
# plan.planDesign.objectives.addObjective(doseObj.EUDMax(roi, 21, 1, weight=50))
# plan.planDesign.objectives.addObjective(doseObj.EUDUniform(roi, 20, 1, weight=50))
Optimize plan#
solver = IntensityModulationOptimizer(method='Scipy_L-BFGS-B', plan=plan, maxiter=50, hardwareAcceleration = None)
doseImage, ps = solver.optimize()
Final dose computation#
mc2.nbPrimaries = 1e6
doseImage = mc2.computeDose(ct, plan)
Plots#
# Compute DVH on resampled contour
roiResampled = resampleImage3D(roi, origin=ct.origin, spacing=scoringSpacing)
target_DVH = DVH(roiResampled, doseImage)
print('D95 = ' + str(target_DVH.D95) + ' Gy')
print('D5 = ' + str(target_DVH.D5) + ' Gy')
print('D5 - D95 = {} Gy'.format(target_DVH.D5 - target_DVH.D95))
bodyResampled = resampleImage3D(body, origin=ct.origin, spacing=scoringSpacing)
body_DVH = DVH(bodyResampled, doseImage)
# center of mass
roi = resampleImage3DOnImage3D(roi, ct)
body = resampleImage3DOnImage3D(body,ct)
COM_coord = roi.centerOfMass
COM_index = roi.getVoxelIndexFromPosition(COM_coord)
Z_coord = COM_index[2]
img_ct = ct.imageArray[:, :, Z_coord].transpose(1, 0)
img_mask = roi.imageArray[:, :, Z_coord].transpose(1, 0)
img_body = body.imageArray[:, :, Z_coord].transpose(1, 0)
img_dose = resampleImage3DOnImage3D(doseImage, ct)
img_dose = img_dose.imageArray[:, :, Z_coord].transpose(1, 0)
#Output path
output_path = 'Output'
if not os.path.exists(output_path):
os.makedirs(output_path)
# Display dose
fig, ax = plt.subplots(1, 2, figsize=(12, 5))
ax[0].imshow(img_ct, cmap='gray')
ax[0].contour(img_body,[0.5],colors='green') # Body
ax[0].contour(img_mask,[0.5],colors='red') # PTV
dose = ax[0].imshow(img_dose, cmap='jet', alpha=.2)
plt.colorbar(dose, ax=ax[0])
ax[1].plot(target_DVH.histogram[0], target_DVH.histogram[1], label=target_DVH.name,color='red')
ax[1].plot(body_DVH.histogram[0], body_DVH.histogram[1], label=body_DVH.name,color='green')
ax[1].set_xlabel("Dose (Gy)")
ax[1].set_ylabel("Volume (%)")
plt.grid(True)
plt.legend()
plt.savefig(os.path.join(output_path, 'SimpleOpti1.png'),format = 'png')
plt.show()
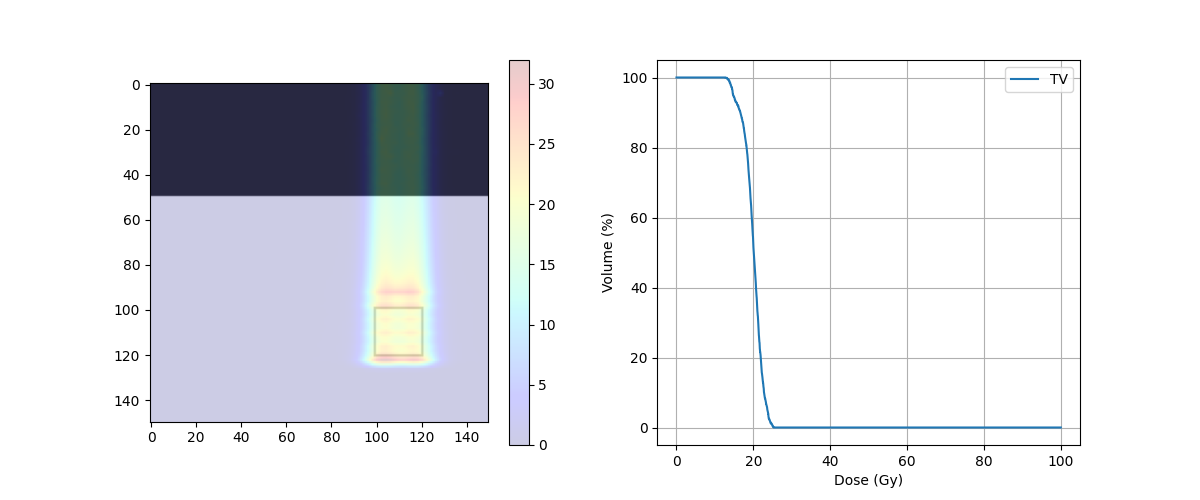
D95 = 18.73779296875 Gy
D5 = 22.6318359375 Gy
D5 - D95 = 3.89404296875 Gy
Total running time of the script: (2 minutes 11.744 seconds)
