Note
Go to the end to download the full example code.
Transform 3D#
author: OpenTPS team
This example demonstrates how to apply a 3D transformation to a synthetic CT image using the OpenTPS library.
running time: ~ 6 minutes
Setting up the environment in google collab#
import sys
if "google.colab" in sys.modules:
from IPython import get_ipython
get_ipython().system('git clone https://gitlab.com/openmcsquare/opentps.git')
get_ipython().system('pip install ./opentps')
import opentps
imports
import copy
import matplotlib.pyplot as plt
from matplotlib.animation import FuncAnimation
import logging
import numpy as np
import math
import os
import the needed opentps.core packages
from opentps.core.data.images import VectorField3D
from opentps.core.data.dynamicData._dynamic3DModel import Dynamic3DModel
from opentps.core.data.dynamicData._dynamic3DSequence import Dynamic3DSequence
from opentps.core.data._transform3D import Transform3D
from opentps.core.processing.imageProcessing.resampler3D import resampleImage3DOnImage3D
from opentps.core.processing.imageProcessing.imageTransform3D import rotateData, translateData
from opentps.core.processing.imageProcessing.resampler3D import resample
from opentps.core.data.images import CTImage
from opentps.core.data.images import ROIMask
logger = logging.getLogger(__name__)
Output path#
output_path = os.path.join(os.getcwd(), 'Output', 'ExampleTransform3D')
if not os.path.exists(output_path):
os.makedirs(output_path)
logger.info('Files will be stored in {}'.format(output_path))
Animation function#
def showModelWithAnimatedFields(model):
for field in model.deformationList:
field.resample(spacing=model.midp.spacing, gridSize=model.midp.gridSize, origin=model.midp.origin)
y_slice = int(model.midp.gridSize[1] / 2)
plt.figure()
fig = plt.gcf()
def updateAnim(imageIndex):
fig.clear()
compX = model.deformationList[imageIndex].velocity.imageArray[:, y_slice, :, 0]
compZ = model.deformationList[imageIndex].velocity.imageArray[:, y_slice, :, 2]
plt.imshow(model.midp.imageArray[:, y_slice, :][::5, ::5], cmap='gray')
plt.quiver(compZ[::5, ::5], compX[::5, ::5], alpha=0.2, color='red', angles='xy', scale_units='xy', scale=5)
anim = FuncAnimation(fig, updateAnim, frames=len(model.deformationList), interval=300)
# anim.save('D:/anim.gif')
plt.show()
Synthetic 4DCT generation function#
def getPhasesPositions(numberOfPhases, minValue, maxValue):
angleList = np.linspace(0, 2 * math.pi, numberOfPhases + 1)[:-1]
cosList = np.cos(angleList)
diff = maxValue - minValue
posList = minValue + diff / 2 + cosList * diff / 2
return posList.astype(np.uint8)
def createSynthetic3DCT(diaphragmPos = 20, targetPos = [50, 100, 35], spacing=[1, 1, 2], returnTumorMask = False):
# GENERATE SYNTHETIC CT IMAGE
# background
im = np.full((170, 170, 100), -1000)
im[20:150, 70:130, :] = 0
# left lung
im[30:70, 80:120, diaphragmPos:] = -800
# right lung
im[100:140, 80:120, diaphragmPos:] = -800
# target
im[targetPos[0]-5:targetPos[0]+5, targetPos[1]-5:targetPos[1]+5, targetPos[2]-5:targetPos[2]+5] = 0
# vertebral column
im[80:90, 95:105, :] = 800
# rib
im[22:26, 90:110, 46:50] = 800
# couch
im[:, 130:135, :] = 100
ct = CTImage(imageArray=im, name='fixed', origin=[0, 0, 0], spacing=spacing)
if returnTumorMask:
mask = np.full((170, 170, 100), 0)
mask[targetPos[0]-5:targetPos[0]+5, targetPos[1]-5:targetPos[1]+5, targetPos[2]-5:targetPos[2]+5] = 1
roi = ROIMask(imageArray=mask, origin=[0, 0, 0], spacing=spacing)
return ct, roi
else:
return ct
def createSynthetic4DCT(numberOfPhases=4, spacing=[1, 1, 2], returnTumorMasks=False, motionNoise=True):
# GENERATE SYNTHETIC 4D INPUT SEQUENCE
CT4D = Dynamic3DSequence()
## For the diaphragm position
diaphMotionAmp = 12
diaphMinPos = 20
diaphPosList = getPhasesPositions(numberOfPhases, diaphMinPos, diaphMinPos+diaphMotionAmp)
if motionNoise:
diaphNoise = [[3, 1],
[6, -1],
[9, -1],
[12, 1],
[15, 1]]
else:
diaphNoise = [[3, 0],
[6, 0],
[9, 0],
[12, 0],
[15, 0]]
for elemIdx in range(len(diaphNoise)):
if diaphNoise[elemIdx][0] <= numberOfPhases - 1:
diaphPosList[diaphNoise[elemIdx][0]] += diaphNoise[elemIdx][1]
## For the target z position
zMotionAmp = int(np.round(diaphMotionAmp * 0.8))
zMinPos = 40
zPosList = getPhasesPositions(numberOfPhases, zMinPos, zMinPos+zMotionAmp)
if motionNoise:
zNoise = [[3, 1],
[6, -1],
[9, -1],
[12, 1],
[15, 1]]
else:
zNoise = [[3, 0],
[6, 0],
[9, 0],
[12, 0],
[15, 0]]
for elemIdx in range(len(zNoise)):
if zNoise[elemIdx][0] <= numberOfPhases - 1:
zPosList[zNoise[elemIdx][0]] += zNoise[elemIdx][1]
## For the target x position
xMotionAmp = 6
xMinPos = 42
xPosList = getPhasesPositions(numberOfPhases, xMinPos, xMinPos+xMotionAmp)
if motionNoise:
xNoise = [[3, 1],
[6, -1],
[9, -1],
[12, 1],
[15, 1]]
else:
xNoise = [[3, 0],
[6, 0],
[9, 0],
[12, 0],
[15, 0]]
for elemIdx in range(len(xNoise)):
if xNoise[elemIdx][0] <= numberOfPhases-1:
xPosList[xNoise[elemIdx][0]] += xNoise[elemIdx][1]
xPosList = np.roll(xPosList, 2)
# print('xPosList', xPosList)
phaseList = []
if returnTumorMasks:
maskList = []
for phaseIndex in range(numberOfPhases):
phase, mask = createSynthetic3DCT(targetPos=[xPosList[phaseIndex], 95, zPosList[phaseIndex]], diaphragmPos=diaphPosList[phaseIndex], spacing=spacing, returnTumorMask=returnTumorMasks)
phaseList.append(phase)
maskList.append(mask)
else:
for phaseIndex in range(numberOfPhases):
phase = createSynthetic3DCT(targetPos=[xPosList[phaseIndex], 95, zPosList[phaseIndex]], diaphragmPos=diaphPosList[phaseIndex], spacing=spacing)
phaseList.append(phase)
CT4D.dyn3DImageList = phaseList
if returnTumorMasks:
return CT4D, maskList
else:
return CT4D
GENERATE SYNTHETIC INPUT IMAGES#
fixed = CTImage()
fixed.imageArray = np.full((20, 20, 20), -1000)
fixed.imageArray[11:16, 5:14, 11:14] = 100.0
moving = copy.copy(fixed)
movingTrans = copy.copy(fixed)
movingRot = copy.copy(fixed)
movingBoth = copy.copy(fixed)
translation = np.array([-5, 0, -2])
rotation = np.array([0, -20, 0])
rotCenter='imgCenter'
Create a transform 3D#
print('Create a transform 3D')
transform3D = Transform3D()
transform3D.initFromTranslationAndRotationVectors(transVec=translation, rotVec=rotation)
transform3D.setCenter(rotCenter)
print('Translation', transform3D.getTranslation())
print('Rotation', transform3D.getRotationAngles(inDegrees=True))
print('moving with transform3D')
moving = transform3D.deformData(moving, outputBox='same')
print('moving translation')
translateData(movingTrans, translationInMM=translation)
print('moving rotation')
rotateData(movingRot, rotAnglesInDeg=rotation, rotCenter=rotCenter, outputBox='same')
# movingRot = resampleImage3DOnImage3D(movingRot, fixedImage=fixed, fillValue=-1000)
print('moving both')
translateData(movingBoth, translationInMM=translation, outputBox='same')
rotateData(movingBoth, rotAnglesInDeg=rotation, rotCenter=rotCenter, outputBox='same')
y_slice = 10
fig, ax = plt.subplots(1, 6)
ax[0].set_title('fixed')
ax[0].imshow(fixed.imageArray[:, y_slice, :])
ax[0].set_xlabel(f"{fixed.origin}\n{fixed.spacing}\n{fixed.gridSize}")
ax[1].set_title('translateData')
ax[1].imshow(movingTrans.imageArray[:, y_slice, :])
ax[1].set_xlabel(f"{movingTrans.origin}\n{movingTrans.spacing}\n{movingTrans.gridSize}")
ax[2].set_title('rotateData')
ax[2].imshow(movingRot.imageArray[:, y_slice, :])
ax[2].set_xlabel(f"{movingRot.origin}\n{movingRot.spacing}\n{movingRot.gridSize}")
ax[3].set_title('both')
ax[3].imshow(movingBoth.imageArray[:, y_slice, :])
ax[3].set_xlabel(f"{movingBoth.origin}\n{movingBoth.spacing}\n{movingBoth.gridSize}")
ax[4].set_title('transform3D')
ax[4].imshow(moving.imageArray[:, y_slice, :])
ax[4].set_xlabel(f"{moving.origin}\n{moving.spacing}\n{moving.gridSize}")
ax[5].set_title('transform3D-both')
ax[5].imshow(moving.imageArray[:, y_slice, :] - movingBoth.imageArray[:, y_slice, :])
plt.savefig(os.path.join(output_path, 'ExampleTransform3D.png'))
plt.show()
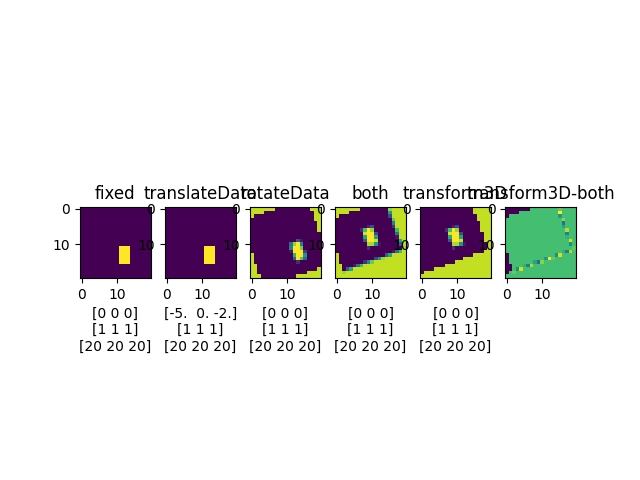
Create a transform 3D
Translation [-5. -0. -2.]
Rotation [ -0. -20. -0.]
moving with transform3D
moving translation
moving rotation
moving both
Create a dynamic model with the transform#
print(' --------------------- start test with model -----------------------------')
CT4D = createSynthetic4DCT(numberOfPhases=4)
# GENERATE MIDP
fixedDynMod = Dynamic3DModel()
fixedDynMod.computeMidPositionImage(CT4D, 0, tryGPU=True)
print(fixedDynMod.midp.origin, fixedDynMod.midp.spacing, fixedDynMod.midp.gridSize)
print('Resample model image')
fixedDynMod = resample(fixedDynMod, gridSize=(80, 50, 50))
print('after resampling', fixedDynMod.midp.origin, fixedDynMod.midp.spacing, fixedDynMod.midp.gridSize)
# option 3
for field in fixedDynMod.deformationList:
print('Resample model field')
field.resample(spacing=fixedDynMod.midp.spacing, gridSize=fixedDynMod.midp.gridSize, origin=fixedDynMod.midp.origin)
print('after resampling', field.origin, field.spacing, field.gridSize)
showModelWithAnimatedFields(fixedDynMod)
movingDynMod = copy.copy(fixedDynMod)
rotateData(movingDynMod, rotAnglesInDeg=rotation, rotCenter=rotCenter, outputBox='same')
showModelWithAnimatedFields(movingDynMod)
--------------------- start test with model -----------------------------
Dynamic 3D Sequence Created with 0 images
[0 0 0] [1 1 2] [170 170 100]
Resample model image
after resampling [0 0 0] [2.125 3.4 4. ] [80 50 50]
Resample model field
after resampling [0 0 0] [2.125 3.4 4. ] [80 50 50]
Resample model field
after resampling [0 0 0] [2.125 3.4 4. ] [80 50 50]
Resample model field
after resampling [0 0 0] [2.125 3.4 4. ] [80 50 50]
Resample model field
after resampling [0 0 0] [2.125 3.4 4. ] [80 50 50]
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/matplotlib/animation.py:908: UserWarning: Animation was deleted without rendering anything. This is most likely not intended. To prevent deletion, assign the Animation to a variable, e.g. `anim`, that exists until you output the Animation using `plt.show()` or `anim.save()`.
warnings.warn(
Generate synthetic input images#
fixed = CTImage()
fixed.imageArray = np.full((20, 20, 20), -1000)
y_slice = 10
pointList = [[15, y_slice, 15], [15, y_slice, 10], [12, y_slice, 12], [10, y_slice, 10]]
for point in pointList:
fixed.imageArray[point[0], point[1], point[2]] = 200
fieldFixed = VectorField3D()
fieldFixed.imageArray = np.zeros((20, 20, 20, 3))
vectorList = [np.array([2, 3, 4]), np.array([0, 3, 4]), np.array([7, 3, 3]), np.array([2, 0, 0])]
for pointIdx in range(len(pointList)):
fieldFixed.imageArray[pointList[pointIdx][0], pointList[pointIdx][1], pointList[pointIdx][2]] = vectorList[
pointIdx]
moving = copy.copy(fixed)
fieldMoving = copy.copy(fieldFixed)
Create a transform 3D#
print('Create a transform 3D')
transform3D = Transform3D()
transform3D.initFromTranslationAndRotationVectors(transVec=translation, rotVec=rotation)
transform3D.setCenter(rotCenter)
print('Translation', transform3D.getTranslation())
print('Rotation', transform3D.getRotationAngles(inDegrees=True))
print('moving with transform3D')
moving = transform3D.deformData(moving, outputBox='same')
fieldMoving = transform3D.deformData(fieldMoving, outputBox='same')
moving = resampleImage3DOnImage3D(moving, fixedImage=fixed, fillValue=-1000)
print('fixed.origin', fixed.origin, 'moving.origin', moving.origin)
fieldMoving = resampleImage3DOnImage3D(fieldMoving, fixedImage=fixed, fillValue=0)
print('fieldFixed.origin', fieldFixed.origin, 'fieldMoving.origin', fieldMoving.origin)
print('ici ', fieldMoving.imageArray[10, y_slice, 10])
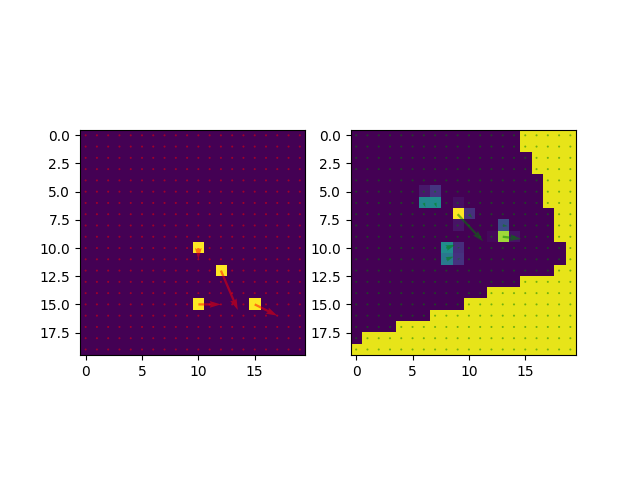
compXFixed = fieldFixed.imageArray[:, y_slice, :, 0]
compZFixed = fieldFixed.imageArray[:, y_slice, :, 2]
compXMoving = fieldMoving.imageArray[:, y_slice, :, 0]
compZMoving = fieldMoving.imageArray[:, y_slice, :, 2]
Create a transform 3D
Translation [-5. -0. -2.]
Rotation [ -0. -20. -0.]
moving with transform3D
fixed.origin [0 0 0] moving.origin [0 0 0]
fieldFixed.origin [0 0 0] fieldMoving.origin [0 0 0]
ici [0. 0. 0.]
Display results#
fig, ax = plt.subplots(1, 2)
ax[0].imshow(fixed.imageArray[:, y_slice, :])
ax[0].quiver(compZFixed, compXFixed, alpha=0.5, color='red', angles='xy', scale_units='xy', scale=2, width=.010)
ax[1].imshow(moving.imageArray[:, y_slice, :])
ax[1].quiver(compZMoving, compXMoving, alpha=0.5, color='green', angles='xy', scale_units='xy', scale=2, width=.010)
plt.show()
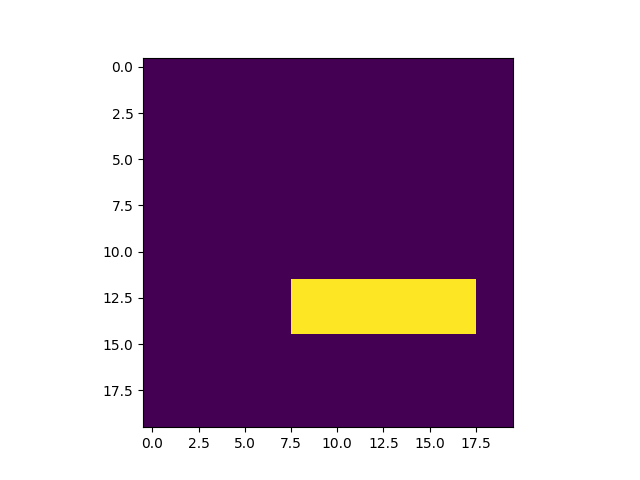
print('start ROIMask test')
fixedMask = ROIMask.fromImage3D(fixed)
fixedMask.imageArray = np.zeros(fixedMask.gridSize).astype(bool)
fixedMask.imageArray[12:15, 8:12, 8:18] = True
plt.figure()
plt.imshow(fixedMask.imageArray[:, y_slice, :])
plt.show()
print(fixedMask.origin, fixedMask.gridSize, fixedMask.spacing, fixedMask.imageArray.dtype)
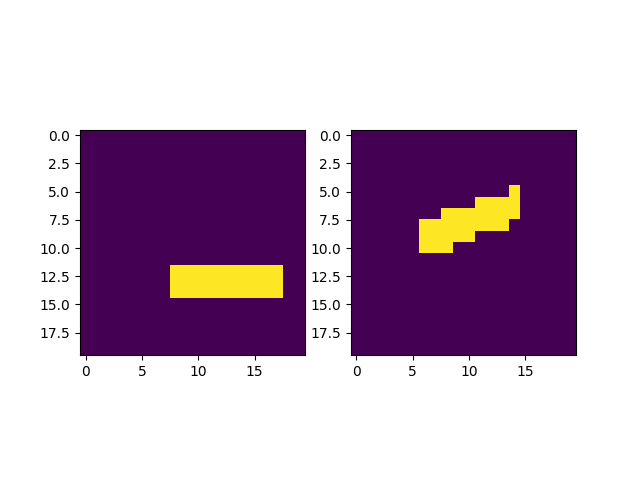
movingMask = copy.copy(fixedMask)
movingMask = transform3D.deformData(movingMask, outputBox='same')
print(movingMask.origin, movingMask.gridSize, movingMask.spacing, movingMask.imageArray.dtype)
plt.figure()
plt.subplot(1, 2, 1)
plt.imshow(fixedMask.imageArray[:, y_slice, :])
plt.subplot(1, 2, 2)
plt.imshow(movingMask.imageArray[:, y_slice, :])
plt.show()
start ROIMask test
[0 0 0] [20 20 20] [1 1 1] bool
[0 0 0] [20 20 20] [1 1 1] bool
Total running time of the script: (0 minutes 16.674 seconds)