Note
Go to the end to download the full example code.
Bound Constraint Optimization#
author: OpenTPS team
In this example, we optimize an ion plan (Protons) using the BoundConstraintsOptimizer function. This function allows optimization with constraints on the Monitor Unit (MU) values of each spot. It helps to stay as close as possible to reality when certain machines cannot accept MU/spot values that are too high or too low.
running time: ~ 15 minutes
Setting up the environment in google collab#
import sys
if "google.colab" in sys.modules:
from IPython import get_ipython
get_ipython().system('git clone https://gitlab.com/openmcsquare/opentps.git')
get_ipython().system('pip install ./opentps')
import opentps
imports
import math
import os
import logging
import numpy as np
from matplotlib import pyplot as plt
import sys
sys.path.append('..')
import the needed opentps.core packages
import opentps.core.processing.planOptimization.objectives.dosimetricObjectives as doseObj
from opentps.core.data.images import CTImage
from opentps.core.data.images import ROIMask
from opentps.core.data.plan import ObjectivesList
from opentps.core.data.plan import ProtonPlanDesign
from opentps.core.data import DVH
from opentps.core.data import Patient
from opentps.core.io import mcsquareIO
from opentps.core.io.scannerReader import readScanner
from opentps.core.io.serializedObjectIO import saveRTPlan, loadRTPlan
from opentps.core.processing.doseCalculation.doseCalculationConfig import DoseCalculationConfig
from opentps.core.processing.doseCalculation.protons.mcsquareDoseCalculator import MCsquareDoseCalculator
from opentps.core.processing.imageProcessing.resampler3D import resampleImage3DOnImage3D, resampleImage3D
from opentps.core.processing.planOptimization.planOptimization import BoundConstraintsOptimizer, IntensityModulationOptimizer
logger = logging.getLogger(__name__)
Output path#
We will create an output folder to store the results of this example
output_path = 'Output'
if not os.path.exists(output_path):
os.makedirs(output_path)
logger.info('Files will be stored in {}'.format(output_path))
Generic CT creation#
we will first create a generic CT of a box fill with water and air
ctCalibration = readScanner(DoseCalculationConfig().scannerFolder)
bdl = mcsquareIO.readBDL(DoseCalculationConfig().bdlFile)
patient = Patient()
patient.name = 'Patient'
ctSize = 150
ct = CTImage()
ct.name = 'CT'
ct.patient = patient
huAir = -1024.
huWater = ctCalibration.convertRSP2HU(1.)
data = huAir * np.ones((ctSize, ctSize, ctSize))
data[:, 50:, :] = huWater
ct.imageArray = data
Region of interest#
we will now create a region of interest wich is a small 3D box of size 20*20*20
roi = ROIMask()
roi.patient = patient
roi.name = 'TV'
roi.color = (255, 0, 0) # red
data = np.zeros((ctSize, ctSize, ctSize)).astype(bool)
data[100:120, 100:120, 100:120] = True
roi.imageArray = data
body = roi.copy()
body.name = 'Body'
body.dilateMask(20)
body.imageArray = np.logical_xor(body.imageArray, roi.imageArray).astype(bool)
Configuration of Mcsquare#
To configure the MCsquare calculator we need to calibrate it with the CT calibration obtained above
mc2 = MCsquareDoseCalculator()
mc2.beamModel = bdl
mc2.ctCalibration = ctCalibration
mc2.nbPrimaries = 5e4
Plan Creation#
# Design plan
beamNames = ["Beam1"]
gantryAngles = [0.]
couchAngles = [0.]
# Load / Generate new plan
plan_file = os.path.join(output_path,"Plan_WaterPhantom_cropped_resampled.tps")
if os.path.isfile(plan_file):
plan = loadRTPlan(plan_file)
logger.info('Plan loaded')
else:
planInit = ProtonPlanDesign()
planInit.ct = ct
planInit.gantryAngles = gantryAngles
planInit.beamNames = beamNames
planInit.couchAngles = couchAngles
planInit.calibration = ctCalibration
planInit.spotSpacing = 5.0
planInit.layerSpacing = 5.0
planInit.targetMargin = 5.0
planInit.setScoringParameters(scoringSpacing=[2, 2, 2], adapt_gridSize_to_new_spacing=True)
# needs to be called after scoringGrid settings but prior to spot placement
planInit.defineTargetMaskAndPrescription(target = roi, targetPrescription = 20.)
plan = planInit.buildPlan() # Spot placement
plan.PlanName = "NewPlan"
beamlets = mc2.computeBeamlets(ct, plan, roi=[roi,body])
plan.planDesign.beamlets = beamlets
beamlets.storeOnFS(os.path.join(output_path, "BeamletMatrix_" + plan.seriesInstanceUID + ".blm"))
saveRTPlan(plan, plan_file)
# Set objectives (attribut is already initialized in planDesign object)
plan.planDesign.objectives.addObjective(doseObj.DMax(body,5, weight=1.0))
plan.planDesign.objectives.addObjective(doseObj.DMax(roi, 21, weight=10.0))
plan.planDesign.objectives.addObjective(doseObj.DMin(roi, 20, weight=20.0))
solver = BoundConstraintsOptimizer(method='Scipy_L-BFGS-B', plan=plan, maxiter=50, bounds=(0.2, 50))
Optimize treatment plan#
doseImage, ps = solver.optimize()
Dose volume histogram#
target_DVH = DVH(roi, doseImage)
body_DVH = DVH(body, doseImage)
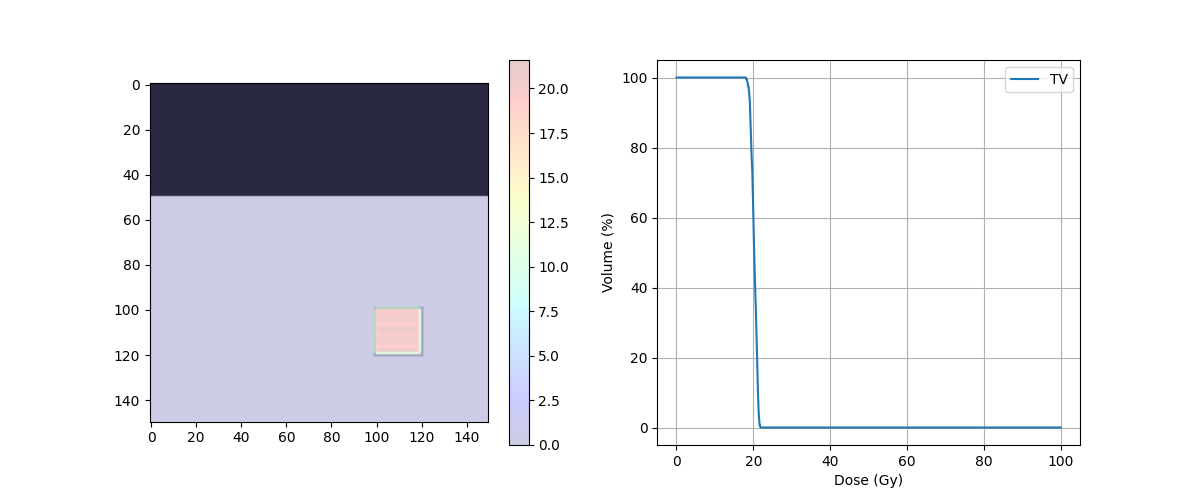
print('D95 = ' + str(target_DVH.D95) + ' Gy')
print('D5 = ' + str(target_DVH.D5) + ' Gy')
print('D5 - D95 = {} Gy'.format(target_DVH.D5 - target_DVH.D95))
D95 = 18.778483072916668 Gy
D5 = 22.38246372767857 Gy
D5 - D95 = 3.6039806547619015 Gy
Center of mass#
Here we look at the part of the 3D CT image where “stuff is happening” by getting the CoM. We use the function resampleImage3DOnImage3D to the same array size for both images.
roi = resampleImage3DOnImage3D(roi, ct)
body = resampleImage3DOnImage3D(body,ct)
COM_coord = roi.centerOfMass
COM_index = roi.getVoxelIndexFromPosition(COM_coord)
Z_coord = COM_index[2]
img_ct = ct.imageArray[:, :, Z_coord].transpose(1, 0)
img_mask = roi.imageArray[:, :, Z_coord].transpose(1, 0)
img_body = body.imageArray[:, :, Z_coord].transpose(1, 0)
img_dose = resampleImage3DOnImage3D(doseImage, ct)
img_dose = img_dose.imageArray[:, :, Z_coord].transpose(1, 0)
Plot of the dose#
fig, ax = plt.subplots(1, 2, figsize=(12, 5))
ax[0].imshow(img_ct, cmap='gray')
ax[0].contour(img_body,[0.5],colors='green') # Body
ax[0].contour(img_mask,[0.5],colors='red') # PTV
dose = ax[0].imshow(img_dose, cmap='jet', alpha=.2)
plt.colorbar(dose, ax=ax[0])
ax[1].plot(target_DVH.histogram[0], target_DVH.histogram[1], label=target_DVH.name,color='red')
ax[1].plot(body_DVH.histogram[0], body_DVH.histogram[1], label=body_DVH.name,color='green')
ax[1].set_xlabel("Dose (Gy)")
ax[1].set_ylabel("Volume (%)")
plt.grid(True)
plt.legend()
plt.savefig(os.path.join(output_path, 'SimpleOpti1.png'),format = 'png')
plt.show()
Total running time of the script: (3 minutes 42.436 seconds)
