Note
Go to the end to download the full example code.
Segmentation#
Author : OpenTPS team
This example will present the basis of segmentation with openTPS core. running time: ~ 5 minutes
Setting up the environment in google collab#
import sys
if "google.colab" in sys.modules:
from IPython import get_ipython
get_ipython().system('git clone https://gitlab.com/openmcsquare/opentps.git')
get_ipython().system('pip install ./opentps')
import opentps
imports
import numpy as np
import matplotlib.pyplot as plt
import logging
import os
import the needed opentps.core packages
from opentps.core.data.images import CTImage
from opentps.core.processing.segmentation.segmentation3D import applyThreshold
from opentps.core.processing.segmentation.segmentationCT import SegmentationCT
from opentps.core.data.images._ctImage import CTImage
from opentps.core.data.images._roiMask import ROIMask
logger = logging.getLogger(__name__)
Output path#
output_path = 'Output'
if not os.path.exists(output_path):
os.makedirs(output_path)
Genrerate synthetic CT image and segment it#
def createSynthetic3DCT(diaphragmPos = 20, targetPos = [50, 100, 35], spacing=[1, 1, 2], returnTumorMask = False):
# GENERATE SYNTHETIC CT IMAGE
# background
im = np.full((170, 170, 100), -1000)
im[20:150, 70:130, :] = 0
# left lung
im[30:70, 80:120, diaphragmPos:] = -800
# right lung
im[100:140, 80:120, diaphragmPos:] = -800
# target
im[targetPos[0]-5:targetPos[0]+5, targetPos[1]-5:targetPos[1]+5, targetPos[2]-5:targetPos[2]+5] = 0
# vertebral column
im[80:90, 95:105, :] = 800
# rib
im[22:26, 90:110, 46:50] = 800
# couch
im[:, 130:135, :] = 100
ct = CTImage(imageArray=im, name='fixed', origin=[0, 0, 0], spacing=spacing)
if returnTumorMask:
mask = np.full((170, 170, 100), 0)
mask[targetPos[0]-5:targetPos[0]+5, targetPos[1]-5:targetPos[1]+5, targetPos[2]-5:targetPos[2]+5] = 1
roi = ROIMask(imageArray=mask, origin=[0, 0, 0], spacing=spacing)
return ct, roi
else:
return ct
# GENERATE SYNTHETIC CT IMAGE
ct = createSynthetic3DCT()
# APPLY THRESHOLD SEGMENTATION
mask = applyThreshold(ct, -750)
# APPLY CT BODY SEGMENTATION
seg = SegmentationCT(ct)
body = seg.segmentBody()
bones = seg.segmentBones()
lungs = seg.segmentLungs()
# CHECK RESULTS
assert (body.imageArray[50,100,80] == True) & (body.imageArray[0,0,0] == False), f"Wrong body segmentation"
assert (bones.imageArray[85,100,50] == True) & (bones.imageArray[85,110,50] == False), f"Wrong bones segmentation"
assert (lungs.imageArray[120,100,35] == True) & (lungs.imageArray[50,100,35] == False), f"Wrong lungs segmentation"
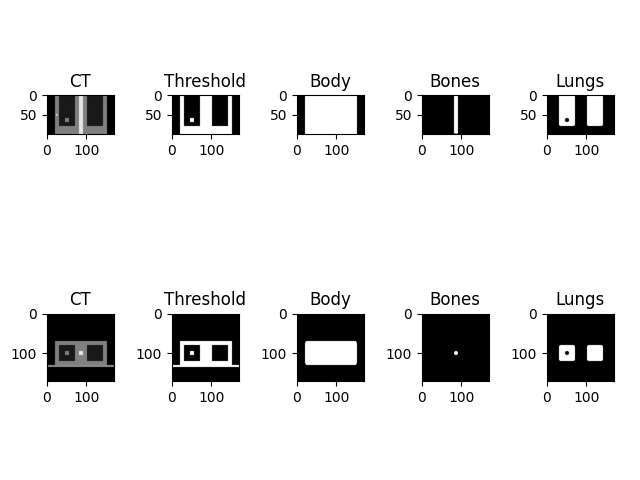
DISPLAY RESULTS#
fig, ax = plt.subplots(2, 5)
fig.tight_layout()
y_slice = 100
z_slice = 35 #round(ct.imageArray.shape[2] / 2) - 1
ax[0,0].imshow(ct.imageArray[:, y_slice, :].T[::-1, ::1], cmap='gray', origin='upper', vmin=-1000, vmax=1000)
ax[0,0].title.set_text('CT')
ax[0,1].imshow(mask.imageArray[:, y_slice, :].T[::-1, ::1], cmap='gray', origin='upper', vmin=0, vmax=1)
ax[0,1].title.set_text('Threshold')
ax[0,2].imshow(body.imageArray[:, y_slice, :].T[::-1, ::1], cmap='gray', origin='upper', vmin=0, vmax=1)
ax[0,2].title.set_text('Body')
ax[0,3].imshow(bones.imageArray[:, y_slice, :].T[::-1, ::1], cmap='gray', origin='upper', vmin=0, vmax=1)
ax[0,3].title.set_text('Bones')
ax[0,4].imshow(lungs.imageArray[:, y_slice, :].T[::-1, ::1], cmap='gray', origin='upper', vmin=0, vmax=1)
ax[0,4].title.set_text('Lungs')
ax[1,0].imshow(ct.imageArray[:, :, z_slice].T[::1, ::1], cmap='gray', origin='upper', vmin=-1000, vmax=1000)
ax[1,0].title.set_text('CT')
ax[1,1].imshow(mask.imageArray[:, :, z_slice].T[::1, ::1], cmap='gray', origin='upper', vmin=0, vmax=1)
ax[1,1].title.set_text('Threshold')
ax[1,2].imshow(body.imageArray[:, :, z_slice].T[::1, ::1], cmap='gray', origin='upper', vmin=0, vmax=1)
ax[1,2].title.set_text('Body')
ax[1,3].imshow(bones.imageArray[:, :, z_slice].T[::1, ::1], cmap='gray', origin='upper', vmin=0, vmax=1)
ax[1,3].title.set_text('Bones')
ax[1,4].imshow(lungs.imageArray[:, :, z_slice].T[::1, ::1], cmap='gray', origin='upper', vmin=0, vmax=1)
ax[1,4].title.set_text('Lungs')
plt.savefig(os.path.join(output_path, 'Example_Segmentation.png'))
print('Segmentation example completed')
plt.show()
Segmentation example completed
Total running time of the script: (0 minutes 11.319 seconds)
