Note
Go to the end to download the full example code.
Proton Plan Creation and Dose Calculation#
author: OpenTPS team
This example demonstrates how to create a proton plan and perform dose calculation using OpenTPS.
running time: ~ 5 minutes
Setting up the environment in google collab#
import sys
if "google.colab" in sys.modules:
from IPython import get_ipython
get_ipython().system('git clone https://gitlab.com/openmcsquare/opentps.git')
get_ipython().system('pip install ./opentps')
import opentps
imports
import os
import logging
import sys
import the needed opentps.core packages
from matplotlib import pyplot as plt
from opentps.core.processing.imageProcessing.resampler3D import resampleImage3DOnImage3D
from opentps.core.processing.planOptimization.tools import evaluateClinical
sys.path.append('..')
import numpy as np
from opentps.core.data.plan import ProtonPlan, RTPlan
from opentps.core.io.scannerReader import readScanner
from opentps.core.io.serializedObjectIO import loadRTPlan, saveRTPlan
from opentps.core.io.dicomIO import readDicomDose, readDicomPlan
from opentps.core.io.dataLoader import readData
from opentps.core.data.CTCalibrations.MCsquareCalibration._mcsquareCTCalibration import MCsquareCTCalibration
from opentps.core.io import mcsquareIO
from opentps.core.data._dvh import DVH
from opentps.core.processing.doseCalculation.doseCalculationConfig import DoseCalculationConfig
from opentps.core.processing.doseCalculation.protons.mcsquareDoseCalculator import MCsquareDoseCalculator
from opentps.core.io.mhdIO import exportImageMHD
from opentps.core.data.plan import PlanProtonBeam
from opentps.core.data.plan import PlanProtonLayer
from opentps.core.data.images import CTImage, DoseImage
from opentps.core.data import RTStruct
from opentps.core.data import Patient
from opentps.core.data.images import ROIMask
from pathlib import Path
logger = logging.getLogger(__name__)
Output path#
output_path = os.getcwd()
logger.info('Files will be stored in {}'.format(output_path))
Create plan from scratch and save it#
plan = ProtonPlan()
plan.appendBeam(PlanProtonBeam())
plan.appendBeam(PlanProtonBeam())
plan.beams[1].gantryAngle = 120.
plan.beams[0].appendLayer(PlanProtonLayer(100))
plan.beams[0].appendLayer(PlanProtonLayer(90))
plan.beams[1].appendLayer(PlanProtonLayer(80))
plan[0].layers[0].appendSpot([-1,0,1], [1,2,3], [0.1,0.2,0.3])
plan[0].layers[1].appendSpot([0,1], [2,3], [0.2,0.3])
plan[1].layers[0].appendSpot(1, 1, 0.5)
# Save plan
saveRTPlan(plan,os.path.join(output_path,'dummy_plan.tps'))
Load plan in OpenTPS format (serialized)
plan2 = loadRTPlan(os.path.join(output_path,'dummy_plan.tps'))
print(plan2[0].layers[1].spotWeights)
print(plan[0].layers[1].spotWeights)
# Load DICOM plan
#dicomPath = os.path.join(Path(os.getcwd()).parent.absolute(),'opentps','testData','Phantom')
#print(dicomPath)
#dataList = readData(dicomPath, maxDepth=1)
#plan3 = [d for d in dataList if isinstance(d, RTPlan)][0]
# or provide path to RTPlan and read it
# plan_path = os.path.join(Path(os.getcwd()).parent.absolute(),'opentps/testData/Phantom/Plan_SmallWaterPhantom_cropped_resampled_optimized.dcm')
# plan3 = readDicomPlan(plan_path)
[0.2 0.3]
[0.2 0.3]
Dose computation from plan#
Choosing default Scanner and BDL
doseCalculator = MCsquareDoseCalculator()
doseCalculator.ctCalibration = readScanner(DoseCalculationConfig().scannerFolder)
doseCalculator.beamModel = mcsquareIO.readBDL(DoseCalculationConfig().bdlFile)
doseCalculator.nbPrimaries = 1e7
Generic example: box of water with spherical target#
# Load CT & contours
# ct = [d for d in dataList if isinstance(d, CTImage)][0]
# struct = [d for d in dataList if isinstance(d, RTStruct)][0]
# target = struct.getContourByName('TV')
# body = struct.getContourByName('Body')
# or create CT and contours
ctSize = 20
ct = CTImage()
ct.name = 'CT'
target = ROIMask()
target.name = 'TV'
target.spacing = ct.spacing
target.color = (255, 0, 0) # red
targetArray = np.zeros((ctSize, ctSize, ctSize)).astype(bool)
radius = 2.5
x0, y0, z0 = (10, 10, 10)
x, y, z = np.mgrid[0:ctSize:1, 0:ctSize:1, 0:ctSize:1]
r = np.sqrt((x - x0) ** 2 + (y - y0) ** 2 + (z - z0) ** 2)
targetArray[r < radius] = True
target.imageArray = targetArray
huAir = -1024.
huWater = doseCalculator.ctCalibration.convertRSP2HU(1.)
ctArray = huAir * np.ones((ctSize, ctSize, ctSize))
ctArray[1:ctSize - 1, 1:ctSize - 1, 1:ctSize - 1] = huWater
ctArray[targetArray>=0.5] = 50
ct.imageArray = ctArray
body = ROIMask()
body.name = 'Body'
body.spacing = ct.spacing
body.color = (0, 0, 255)
bodyArray = np.zeros((ctSize, ctSize, ctSize)).astype(bool)
bodyArray[1:ctSize- 1, 1:ctSize - 1, 1:ctSize - 1] = True
body.imageArray = bodyArray
# MCsquare simulation
doseImage = doseCalculator.computeDose(ct, plan)
# or Load dicom dose
#doseImage = [d for d in dataList if isinstance(d, DoseImage)][0]
# or
#dcm_dose_file = os.path.join(output_path, "Dose_SmallWaterPhantom_resampled_optimized.dcm")
#doseImage = readDicomDose(dcm_dose_file)
# Export dose
#output_path = os.getcwd()
#exportImageMHD(output_path, doseImage)
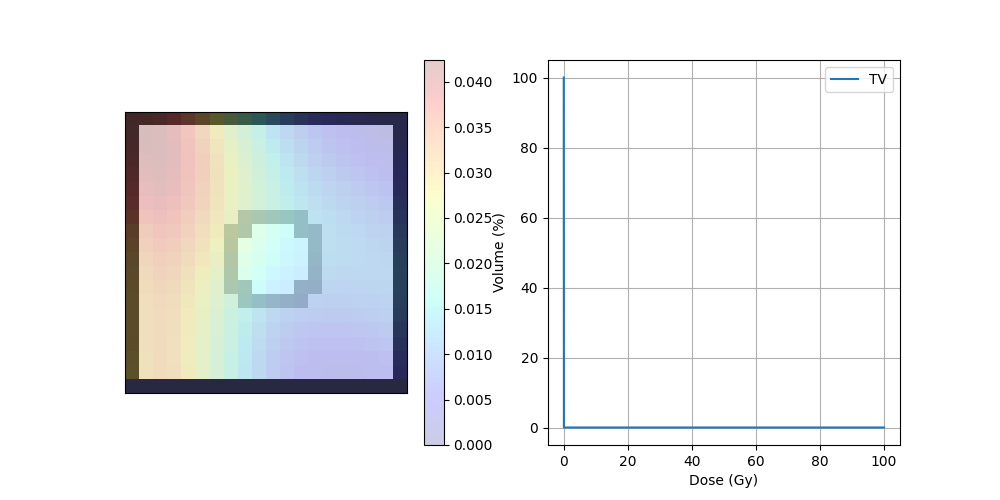
DVH#
dvh = DVH(target, doseImage)
print("D95",dvh._D95)
print("D5",dvh._D5)
print("Dmax",dvh._Dmax)
print("Dmin",dvh._Dmin)
D95 0.013584234775641024
D5 0.05029296874999999
Dmax 0.02884376954615371
Dmin 0.006799805725132493
Plot dose#
target = resampleImage3DOnImage3D(target, ct)
COM_coord = target.centerOfMass
COM_index = target.getVoxelIndexFromPosition(COM_coord)
Z_coord = COM_index[2]
img_ct = ct.imageArray[:, :, Z_coord].transpose(1, 0)
contourTargetMask = target.getBinaryContourMask()
img_mask = contourTargetMask.imageArray[:, :, Z_coord].transpose(1, 0)
img_dose = resampleImage3DOnImage3D(doseImage, ct)
img_dose = img_dose.imageArray[:, :, Z_coord].transpose(1, 0)
Display dose#
fig, ax = plt.subplots(1, 2, figsize=(10, 5))
ax[0].axes.get_xaxis().set_visible(False)
ax[0].axes.get_yaxis().set_visible(False)
ax[0].imshow(img_ct, cmap='gray')
ax[0].imshow(img_mask, alpha=.2, cmap='binary') # PTV
dose = ax[0].imshow(img_dose, cmap='jet', alpha=.2)
plt.colorbar(dose, ax=ax[0])
ax[1].plot(dvh.histogram[0], dvh.histogram[1], label=dvh.name)
ax[1].set_xlabel("Dose (Gy)")
ax[1].set_ylabel("Volume (%)")
ax[1].grid(True)
ax[1].legend()
plt.show()
Total running time of the script: (0 minutes 13.323 seconds)
