Note
Go to the end to download the full example code.
Robust Proton Plan Optimization#
author: OpenTPS team
In this example, we create and optimize a robust proton plan. The setup and range errors are configurable.
running time: ~ 20 minutes
Setting up the environment in google collab#
import sys
if "google.colab" in sys.modules:
from IPython import get_ipython
get_ipython().system('git clone https://gitlab.com/openmcsquare/opentps.git')
get_ipython().system('pip install ./opentps')
import opentps
imports
import os
import logging
import numpy as np
from matplotlib import pyplot as plt
import sys
sys.path.append('..')
import the needed opentps.core packages
import opentps.core.processing.planOptimization.objectives.dosimetricObjectives as doseObj
from opentps.core.data.images import CTImage
from opentps.core.data.images import ROIMask
from opentps.core.data.plan._protonPlanDesign import ProtonPlanDesign
from opentps.core.data.plan import RobustnessProton
from opentps.core.data import DVH
from opentps.core.data import Patient
from opentps.core.io import mcsquareIO
from opentps.core.io.scannerReader import readScanner
from opentps.core.io.serializedObjectIO import loadRTPlan, saveRTPlan
from opentps.core.processing.doseCalculation.doseCalculationConfig import DoseCalculationConfig
from opentps.core.processing.doseCalculation.protons.mcsquareDoseCalculator import MCsquareDoseCalculator
from opentps.core.processing.imageProcessing.resampler3D import resampleImage3DOnImage3D
from opentps.core.processing.planOptimization.planOptimization import IntensityModulationOptimizer
logger = logging.getLogger(__name__)
Output path#
output_path = os.path.join(os.getcwd(), 'Proton_Robust_Output_Example')
if not os.path.exists(output_path):
os.makedirs(output_path)
logger.info('Files will be stored in {}'.format(output_path))
CT calibration and BDL#
ctCalibration = readScanner(DoseCalculationConfig().scannerFolder)
bdl = mcsquareIO.readBDL(DoseCalculationConfig().bdlFile)
Create synthetic CT and ROI#
patient = Patient()
patient.name = 'Patient'
ctSize = 150
ct = CTImage()
ct.name = 'CT'
ct.patient = patient
huAir = -1024.
huWater = ctCalibration.convertRSP2HU(1.)
data = huAir * np.ones((ctSize, ctSize, ctSize))
data[:, 50:, :] = huWater
ct.imageArray = data
roi = ROIMask()
roi.patient = patient
roi.name = 'TV'
roi.color = (255, 0, 0) # red
data = np.zeros((ctSize, ctSize, ctSize)).astype(bool)
data[100:120, 100:120, 100:120] = True
roi.imageArray = data
body = roi.copy()
body.name = 'Body'
body.dilateMask(20)
body.imageArray = np.logical_xor(body.imageArray, roi.imageArray).astype(bool)
Design plan#
beamNames = ["Beam1"]
gantryAngles = [0.]
couchAngles = [0.]
Create output folder#
if not os.path.isdir(output_path):
os.mkdir(output_path)
Configure MCsquare#
mc2 = MCsquareDoseCalculator()
mc2.beamModel = bdl
mc2.nbPrimaries = 1e3
mc2.ctCalibration = ctCalibration
Load / Generate new plan#
plan_file = os.path.join(output_path, "RobustPlan_notCropped.tps")
if os.path.isfile(plan_file):
plan = loadRTPlan(plan_file)
logger.info('Plan loaded')
else:
planDesign = ProtonPlanDesign()
planDesign.ct = ct
planDesign.gantryAngles = gantryAngles
planDesign.beamNames = beamNames
planDesign.couchAngles = couchAngles
planDesign.calibration = ctCalibration
# Robustness settings
planDesign.robustness = RobustnessProton()
planDesign.robustness.setupSystematicError = [1.6, 1.6, 1.6] # mm
planDesign.robustness.setupRandomError = [0.0, 0.0, 0.0] # mm (sigma)
planDesign.robustness.rangeSystematicError = 5.0 # %
# Regular scenario sampling
planDesign.robustness.selectionStrategy = planDesign.robustness.Strategies.REDUCED_SET
# All scenarios (includes diagonals on sphere)
# planDesign.robustness.selectionStrategy = planDesign.robustness.Strategies.ALL
# Random scenario sampling
# planDesign.robustness.selectionStrategy = planDesign.robustness.Strategies.RANDOM
planDesign.robustness.numScenarios = 5 # specify how many random scenarios to simulate, default = 100
planDesign.spotSpacing = 7.0
planDesign.layerSpacing = 6.0
planDesign.targetMargin = max(planDesign.spotSpacing, planDesign.layerSpacing) + max(planDesign.robustness.setupSystematicError)
# scoringGridSize = [int(math.floor(i / j * k)) for i, j, k in zip(ct.gridSize, scoringSpacing, ct.spacing)]
# planDesign.objectives.setScoringParameters(ct, scoringGridSize, scoringSpacing)
planDesign.defineTargetMaskAndPrescription(target = roi, targetPrescription = 20.) # needs to be called prior spot placement
plan = planDesign.buildPlan() # Spot placement
plan.PlanName = "RobustPlan"
nominal, scenarios = mc2.computeRobustScenarioBeamlets(ct, plan, roi=[roi,body], storePath=output_path)
plan.planDesign.beamlets = nominal
plan.planDesign.robustness.scenarios = scenarios
plan.planDesign.robustness.numScenarios = len(scenarios)
#saveRTPlan(plan, plan_file)
saveRTPlan(plan, plan_file)
Set objectives (attribut is already initialized in planDesign object)#
plan.planDesign.objectives.addObjective(doseObj.DMax(body,5, weight=1.0))
plan.planDesign.objectives.addObjective(doseObj.DMax(roi, 21, weight=10.0))
plan.planDesign.objectives.addObjective(doseObj.DMin(roi, 20, weight=20.0))
solver = IntensityModulationOptimizer(method='Scipy_L-BFGS-B', plan=plan, maxiter=50)
Optimize treatment plan#
doseImage, ps = solver.optimize()
plan_file = os.path.join(output_path, "Plan_Proton_WaterPhantom_cropped_optimized.tps")
saveRTPlan(plan, plan_file, unloadBeamlets=False)
mc2.nbPrimaries = 1e6
doseImage = mc2.computeDose(ct, plan)
Compute DVH#
target_DVH = DVH(roi, doseImage)
body_DVH = DVH(body, doseImage)
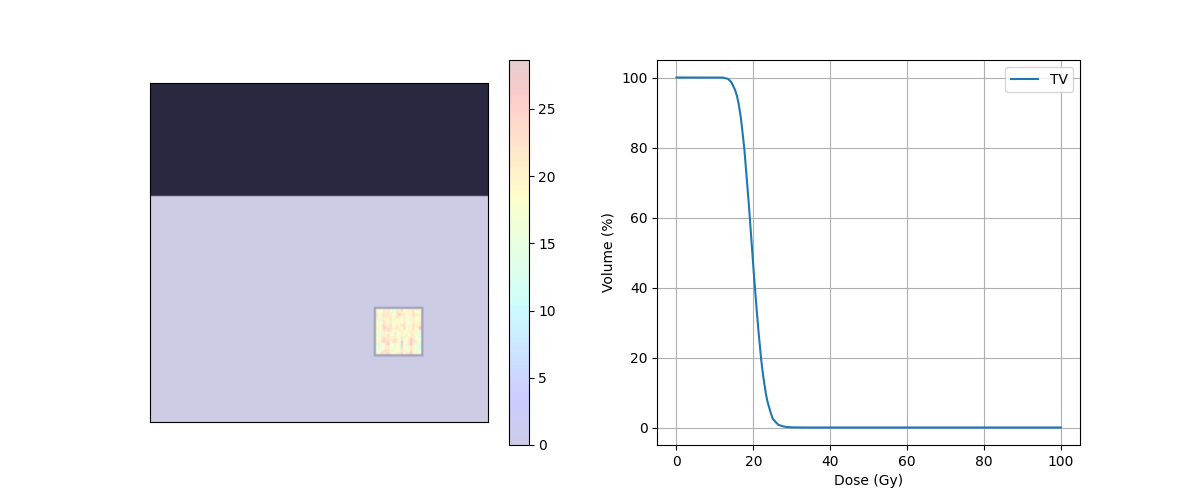
print('D95 = ' + str(target_DVH.D95) + ' Gy')
print('D5 = ' + str(target_DVH.D5) + ' Gy')
print('D5 - D95 = {} Gy'.format(target_DVH.D5 - target_DVH.D95))
D95 = 17.892733487215907 Gy
D5 = 23.529488699776785 Gy
D5 - D95 = 5.636755212560878 Gy
Center of mass#
roi = resampleImage3DOnImage3D(roi, ct)
body = resampleImage3DOnImage3D(body,ct)
COM_coord = roi.centerOfMass
COM_index = roi.getVoxelIndexFromPosition(COM_coord)
Z_coord = COM_index[2]
img_ct = ct.imageArray[:, :, Z_coord].transpose(1, 0)
img_mask = roi.imageArray[:, :, Z_coord].transpose(1, 0)
img_body = body.imageArray[:, :, Z_coord].transpose(1, 0)
img_dose = resampleImage3DOnImage3D(doseImage, ct)
img_dose = img_dose.imageArray[:, :, Z_coord].transpose(1, 0)
Display dose#
fig, ax = plt.subplots(1, 2, figsize=(12, 5))
ax[0].axes.get_xaxis().set_visible(False)
ax[0].axes.get_yaxis().set_visible(False)
ax[0].imshow(img_ct, cmap='gray')
ax[0].contour(img_body,[0.5],colors='green') # Body
ax[0].contour(img_mask,[0.5],colors='red') # PTV
dose = ax[0].imshow(img_dose, cmap='jet', alpha=.2)
plt.colorbar(dose, ax=ax[0])
ax[1].plot(target_DVH.histogram[0], target_DVH.histogram[1], label=target_DVH.name,color='red')
ax[1].plot(body_DVH.histogram[0], body_DVH.histogram[1], label=body_DVH.name,color='green')
ax[1].set_xlabel("Dose (Gy)")
ax[1].set_ylabel("Volume (%)")
plt.grid(True)
plt.legend()
plt.savefig(os.path.join(output_path, 'Dose_RobustOptimizationProtons.png'))
plt.show()
Total running time of the script: (5 minutes 39.487 seconds)
