Note
Go to the end to download the full example code.
4D Proton Optimization#
author: OpenTPS team
This example shows how to create and optimize a 4D proton plan using OpenTPS.
running time: ~ 1 hours
Setting up the environment in google collab#
import sys
if "google.colab" in sys.modules:
from IPython import get_ipython
get_ipython().system('git clone https://gitlab.com/openmcsquare/opentps.git')
get_ipython().system('pip install ./opentps')
import opentps
imports
import os
import logging
import numpy as np
from matplotlib import pyplot as plt
import sys
import pydicom
import datetime
sys.path.append('..')
import the needed opentps.core packages
from opentps.core.data.images import CTImage
from opentps.core.data.images import ROIMask
from opentps.core.data.plan._protonPlanDesign import ProtonPlanDesign
from opentps.core.data import DVH
from opentps.core.data import Patient
from opentps.core.io import mcsquareIO
from opentps.core.io.dataLoader import readData
from opentps.core.io.scannerReader import readScanner
from opentps.core.io.serializedObjectIO import loadRTPlan, saveRTPlan
from opentps.core.processing.doseCalculation.doseCalculationConfig import DoseCalculationConfig
from opentps.core.processing.doseCalculation.protons.mcsquareDoseCalculator import MCsquareDoseCalculator
from opentps.core.processing.imageProcessing.resampler3D import resampleImage3DOnImage3D
from opentps.core.processing.planOptimization.planOptimization import IntensityModulationOptimizer
from opentps.core.processing.imageProcessing.resampler3D import resampleImage3DOnImage3D
from opentps.core.io.dicomIO import writeRTDose, readDicomDose
import opentps.core.processing.planOptimization.objectives.dosimetricObjectives as doseObj
logger = logging.getLogger(__name__)
Output path#
output_path = os.path.join(os.getcwd(), 'Exemple_Robust4DOptimization')
if not os.path.exists(output_path):
os.makedirs(output_path)
logger.info('Files will be stored in {}'.format(output_path))
CT calibration and BDL#
ctCalibration = readScanner(DoseCalculationConfig().scannerFolder)
bdl = mcsquareIO.readBDL(DoseCalculationConfig().bdlFile)
CT and ROI creation#
Generic example: 4DCT composed of 3 CTs : 2 phases and the MidP. The anatomy consists of a square target moving vertically, with an organ at risk and soft tissue (muscle) in front of it.
CT4D = []
ROI4D = []
for i in range(0, 3):
# ++++Don't delete UIDs to build the simple study+++++++++++++++++++
studyInstanceUID = pydicom.uid.generate_uid()
ctSeriesInstanceUID = pydicom.uid.generate_uid()
frameOfReferenceUID = pydicom.uid.generate_uid()
# structSeriesInstanceUID = pydicom.uid.generate_uid()
dt = datetime.datetime.now()
#++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# CT
patient = Patient()
patient.name = f'Miro_OpenTPS_4DCT'
Patient.id = f'12082024'
Patient.birthDate = dt.strftime('%Y%m%d')
patient.sex = ""
ctSize = 150
ct = CTImage(seriesInstanceUID=ctSeriesInstanceUID, frameOfReferenceUID=frameOfReferenceUID)
ct.name = f'CT_Phase_{i}'
ct.patient = patient
ct.studyInstanceUID = studyInstanceUID
huWater = 50
huTarget = 100
huMuscle = 200
data = huWater * np.ones((ctSize, ctSize, ctSize))
# Muscle
data[100:140, 20:130, 55:95] = huMuscle
# OAR
data[70:80, 70:80, 65:85] = huTarget
# TargetVolume
if i == 0 :
data[25:45, 70:100, 65:85] = huTarget
if i == 1 :
data[25:45, 60:90, 65:85] = huTarget
if i == 2 :
data[25:45, 50:80, 65:85] = huTarget
ct.imageArray = data
# writeDicomCT(ct, output_path)
#---------------------ROI
ROI = []
# TargetVolume
TV = ROIMask()
TV.patient = patient
TV.name = 'TV'
TV.color = (255, 0, 0) # red
data = np.zeros((ctSize, ctSize, ctSize)).astype(bool)
if i == 0 :
data[25:45, 70:100, 65:85] = True
if i == 1 :
data[25:45, 60:90, 65:85] = True
if i == 2 :
data[25:45, 50:80, 65:85] = True
TV.imageArray = data
ROI.append(TV)
# Muscle
Muscle = ROIMask()
Muscle.patient = patient
Muscle.name = 'Muscle'
Muscle.color = (150, 0, 0)
data = np.zeros((ctSize, ctSize, ctSize)).astype(bool)
data[100:140, 20:130, 55:95] = True
Muscle.imageArray = data
ROI.append(Muscle)
# OAR
OAR = ROIMask()
OAR.patient = patient
OAR.name = 'OAR'
OAR.color = (100, 0, 0)
data = np.zeros((ctSize, ctSize, ctSize)).astype(bool)
data[70:80, 70:80, 65:85] = True
OAR.imageArray = data
ROI.append(OAR)
# Body
BODY = ROIMask()
BODY.patient = patient
BODY.name = 'Body'
BODY.color = (100, 0, 0)
data = np.ones((ctSize, ctSize, ctSize)).astype(bool)
data[np.where(OAR.imageArray)] = False
data[np.where(Muscle.imageArray)] = False
data[np.where(TV.imageArray)] = False
BODY.imageArray = data
ROI.append(BODY)
CT4D.append(ct)
ROI4D.append(ROI)
RefCT = CT4D[1]
RefTV = ROI4D[1][0]
RefOAR = ROI4D[1][2]
RefBody = ROI4D[1][3]
Design plan#
beamNames = ["Beam1"]
gantryAngles = [90.]
couchAngles = [0.]
Configure MCsquare#
mc2 = MCsquareDoseCalculator()
mc2.beamModel = bdl
mc2.nbPrimaries = 1e3
mc2.ctCalibration = ctCalibration
Load / Generate new plan#
plan_file = os.path.join(output_path, f"RobustPlan_4D.tps")
if os.path.isfile(plan_file):
plan = loadRTPlan(plan_file)
logger.info('Plan loaded')
else:
planDesign = ProtonPlanDesign()
planDesign.ct = RefCT # Here, it's the MidP
planDesign.targetMask = RefTV
planDesign.gantryAngles = gantryAngles
planDesign.beamNames = beamNames
planDesign.couchAngles = couchAngles
planDesign.calibration = ctCalibration
# Robustness settings
planDesign.robustness.setupSystematicError = [1.6, 1.6, 1.6] # mm (sigma)
planDesign.robustness.setupRandomError = [0.0, 0.0, 0.0] # mm (sigma)
planDesign.robustness.rangeSystematicError = 3.0 # %
# 4D Evaluation mode
planDesign.robustness.Mode4D = planDesign.robustness.Mode4D.MCsquareAccumulation # Or MCsquareSystematic
planDesign.robustness.selectionStrategy = planDesign.robustness.Strategies.REDUCED_SET # RANDOM not available for MCsquareSystematic
# planDesign.robustness.selectionStrategy = planDesign.robustness.Strategies.ALL (includes diagonals on sphere)
# planDesign.robustness.selectionStrategy = planDesign.robustness.Strategies.RANDOM
planDesign.robustness.numScenarios = 50 # Specify how many random scenarios to simulate, default = 100
# # 4D settings : only for the mode MCsquareAccumulation with the RANDOM strategie
# planDesign.robustness.Create4DCTfromRef = True
# planDesign.robustness.SystematicAmplitudeError = 5.0 # % # Only with RANDOM strategie
# planDesign.robustness.RandomAmplitudeError = 5.0 # %
# planDesign.robustness.Dynamic_delivery = True
# planDesign.robustness.SystematicPeriodError = 5.0 # % # Spot timing required. If not, we calculate them with SimpleBeamDeliveryTimings()
# planDesign.robustness.RandomPeriodError = 5.0 # %
# planDesign.robustness.Breathing_period = 1 # x100% # default value
planDesign.spotSpacing = 10.0
planDesign.layerSpacing = 10.0
planDesign.targetMargin = 7 # Enough to encompass target motion
planDesign.defineTargetMaskAndPrescription(target = RefTV, targetPrescription = 60.)
plan = planDesign.buildPlan()
plan.rtPlanName = f"RobustPlan_4D"
# refIndex :
# ACCUMULATED -> Index of the Image in the 4DCT one wish we will accumulate the dose.
## SYSTEMATIC -> Index of the Image in the 4DCT who will be used as the nominal. So the one closer to the MidP. Or the Midp.
nominal, scenarios = mc2.compute4DRobustScenarioBeamlets(CT4D, plan, refIndex=1, roi=ROI4D, storePath=output_path)
plan.planDesign.beamlets = nominal
plan.planDesign.robustness.scenarios = scenarios
plan.planDesign.robustness.numScenarios = len(scenarios)
saveRTPlan(plan, plan_file)
Dynamic 3D Sequence Created with 3 images
CT_Phase_0
CT_Phase_1
CT_Phase_2
Set objectives#
plan.planDesign.objectives.addObjective(doseObj.DMax(RefTV,63, weight=100.0, robust=True))
plan.planDesign.objectives.addObjective(doseObj.DMin(RefTV,60, weight=100.0, robust=True))
plan.planDesign.objectives.addObjective(doseObj.DMax(RefOAR, 40, weight=80.0))
plan.planDesign.objectives.addObjective(doseObj.DMax(RefBody, 40, weight=80.0))
Optimize treatment plan#
DoseFile = 'DoseRobustPlan4D'
Dose_file = os.path.join(output_path, DoseFile + '.dcm')
if os.path.isfile(Dose_file):
doseImage = readDicomDose(Dose_file)
print('Dose imported')
else :
plan.planDesign.ROI_cropping = False
solver = IntensityModulationOptimizer(method='Scipy_L-BFGS-B', plan=plan, maxiter=150)
# Optimize treatment plan
doseImage, ps = solver.optimize()
saveRTPlan(plan, os.path.join(output_path, "RobustPlan_4D_weighted.tps"))
writeRTDose(doseImage, output_path, DoseFile)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/pydicom/dataset.py:2710: UserWarning: Camel case attribute 'SoftwareVersion' used which is not in the element keyword data dictionary
warn_and_log(msg)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/pydicom/dataset.py:2710: UserWarning: Camel case attribute 'OperatorName' used which is not in the element keyword data dictionary
warn_and_log(msg)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/pydicom/dataset.py:2710: UserWarning: Camel case attribute 'Width' used which is not in the element keyword data dictionary
warn_and_log(msg)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/pydicom/valuerep.py:440: UserWarning: A value of type 'int64' cannot be assigned to a tag with VR US.
warn_and_log(msg)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/pydicom/dataset.py:2710: UserWarning: Camel case attribute 'Height' used which is not in the element keyword data dictionary
warn_and_log(msg)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/pydicom/dataset.py:2710: UserWarning: Camel case attribute 'ColorType' used which is not in the element keyword data dictionary
warn_and_log(msg)
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/pydicom/dataset.py:2710: UserWarning: Camel case attribute 'BitDepth' used which is not in the element keyword data dictionary
warn_and_log(msg)
Display results#
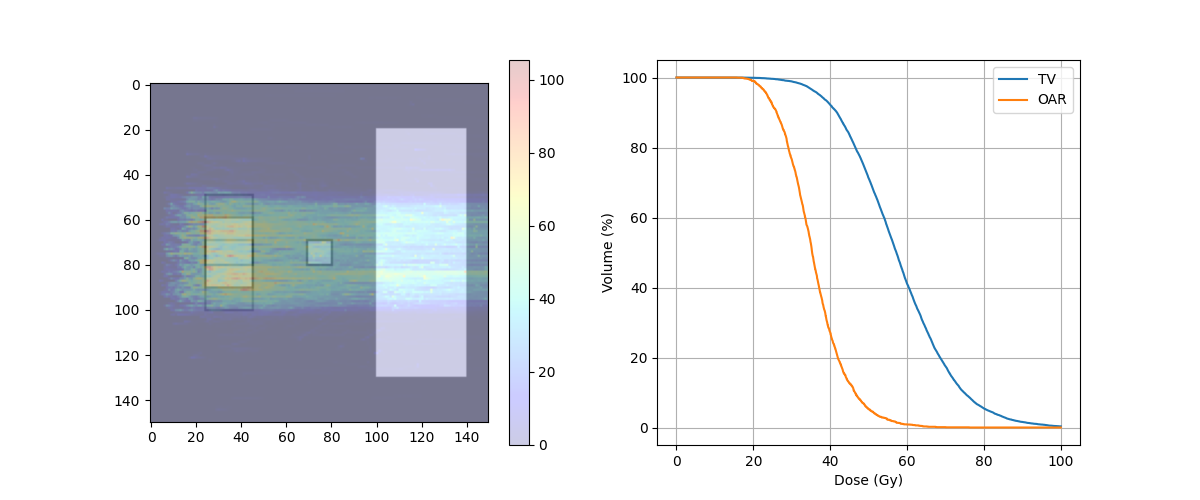
target_DVH = DVH(RefTV, doseImage)
print('TV -> D95 = ' + str(target_DVH.D95) + ' Gy')
print('TV -> D5 = ' + str(target_DVH.D5) + ' Gy')
print('TV -> D5 - D95 = {} Gy'.format(target_DVH.D5 - target_DVH.D95))
oar = resampleImage3DOnImage3D(RefOAR, ct)
oar_DVH = DVH(oar, doseImage)
print('OAR -> D95 = ' + str(oar_DVH.D95) + ' Gy')
print('OAR -> DMAX = ' + str(oar_DVH.Dmax) + ' Gy')
Body = resampleImage3DOnImage3D(RefBody, ct)
Body_DVH = DVH(Body, doseImage)
print('Body -> D95 = ' + str(Body_DVH.D95) + ' Gy')
print('Body -> DMAX = ' + str(Body_DVH.Dmax) + ' Gy')
TV -> D95 = 37.87841796875 Gy
TV -> D5 = 80.48909505208334 Gy
TV -> D5 - D95 = 42.61067708333334 Gy
OAR -> D95 = 25.439453125 Gy
OAR -> DMAX = 109.91547 Gy
Body -> D95 = 0.013658148569185092 Gy
Body -> DMAX = 128.60095 Gy
center of mass#
RefTV = resampleImage3DOnImage3D(RefTV, RefCT)
COM_coord = RefTV.centerOfMass
COM_index = RefTV.getVoxelIndexFromPosition(COM_coord)
Z_coord = COM_index[2]
img_ct = RefCT.imageArray[:, :, Z_coord].transpose(1, 0)
contourTargetMask = RefTV.getBinaryContourMask()
img_mask = contourTargetMask.imageArray[:, :, Z_coord].transpose(1, 0)
img_dose = resampleImage3DOnImage3D(doseImage, RefCT)
img_dose = img_dose.imageArray[:, :, Z_coord].transpose(1, 0)
contourTargetMask0 = ROI4D[0][0].getBinaryContourMask()
img_maskP1 = contourTargetMask0.imageArray[:, :, Z_coord].transpose(1, 0)
contourTargetMask2 = ROI4D[2][0].getBinaryContourMask()
img_maskP2 = contourTargetMask2.imageArray[:, :, Z_coord].transpose(1, 0)
contourOAR = RefOAR.getBinaryContourMask()
img_OAR = contourOAR.imageArray[:, :, Z_coord].transpose(1, 0)
Display dose#
fig, ax = plt.subplots(1, 2, figsize=(12, 5))
ax[0].imshow(img_ct, cmap='gray')
ax[0].imshow(img_mask, alpha=.2, cmap='binary') # PTV
ax[0].imshow(img_maskP1, alpha=.2, cmap='binary')
ax[0].imshow(img_maskP2, alpha=.2, cmap='binary')
ax[0].imshow(img_OAR, alpha=.2, cmap='binary')
dose = ax[0].imshow(img_dose, cmap='jet', alpha=.2)
plt.colorbar(dose, ax=ax[0])
ax[1].plot(target_DVH.histogram[0], target_DVH.histogram[1], label=target_DVH.name)
ax[1].plot(oar_DVH.histogram[0], oar_DVH.histogram[1], label=oar_DVH.name)
ax[1].set_xlabel("Dose (Gy)")
ax[1].set_ylabel("Volume (%)")
plt.grid(True)
plt.legend()
plt.savefig(f'{output_path}/DoseRobustOptimization_4D.png', format = 'png')
plt.show()
Total running time of the script: (11 minutes 3.827 seconds)
